Rüveyda Yilmaz
Diffusion-based Sinogram Interpolation for Limited Angle PET
Nov 12, 2025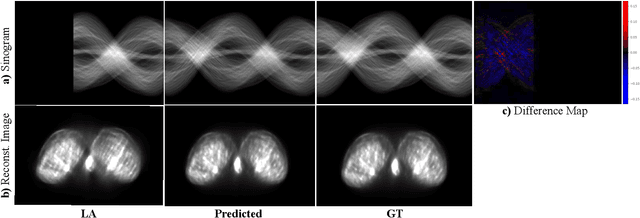
Abstract:Accurate PET imaging increasingly requires methods that support unconstrained detector layouts from walk-through designs to long-axial rings where gaps and open sides lead to severely undersampled sinograms. Instead of constraining the hardware to form complete cylinders, we propose treating the missing lines-of-responses as a learnable prior. Data-driven approaches, particularly generative models, offer a promising pathway to recover this missing information. In this work, we explore the use of conditional diffusion models to interpolate sparsely sampled sinograms, paving the way for novel, cost-efficient, and patient-friendly PET geometries in real clinical settings.
CellStyle: Improved Zero-Shot Cell Segmentation via Style Transfer
Mar 11, 2025Abstract:Cell microscopy data are abundant; however, corresponding segmentation annotations remain scarce. Moreover, variations in cell types, imaging devices, and staining techniques introduce significant domain gaps between datasets. As a result, even large, pretrained segmentation models trained on diverse datasets (source datasets) struggle to generalize to unseen datasets (target datasets). To overcome this generalization problem, we propose CellStyle, which improves the segmentation quality of such models without requiring labels for the target dataset, thereby enabling zero-shot adaptation. CellStyle transfers the attributes of an unannotated target dataset, such as texture, color, and noise, to the annotated source dataset. This transfer is performed while preserving the cell shapes of the source images, ensuring that the existing source annotations can still be used while maintaining the visual characteristics of the target dataset. The styled synthetic images with the existing annotations enable the finetuning of a generalist segmentation model for application to the unannotated target data. We demonstrate that CellStyle significantly improves zero-shot cell segmentation performance across diverse datasets by finetuning multiple segmentation models on the style-transferred data. The code will be made publicly available.
A Pragmatic Note on Evaluating Generative Models with Fréchet Inception Distance for Retinal Image Synthesis
Feb 26, 2025Abstract:Fr\'echet Inception Distance (FID), computed with an ImageNet pretrained Inception-v3 network, is widely used as a state-of-the-art evaluation metric for generative models. It assumes that feature vectors from Inception-v3 follow a multivariate Gaussian distribution and calculates the 2-Wasserstein distance based on their means and covariances. While FID effectively measures how closely synthetic data match real data in many image synthesis tasks, the primary goal in biomedical generative models is often to enrich training datasets ideally with corresponding annotations. For this purpose, the gold standard for evaluating generative models is to incorporate synthetic data into downstream task training, such as classification and segmentation, to pragmatically assess its performance. In this paper, we examine cases from retinal imaging modalities, including color fundus photography and optical coherence tomography, where FID and its related metrics misalign with task-specific evaluation goals in classification and segmentation. We highlight the limitations of using various metrics, represented by FID and its variants, as evaluation criteria for these applications and address their potential caveats in broader biomedical imaging modalities and downstream tasks.
Cascaded Diffusion Models for 2D and 3D Microscopy Image Synthesis to Enhance Cell Segmentation
Nov 19, 2024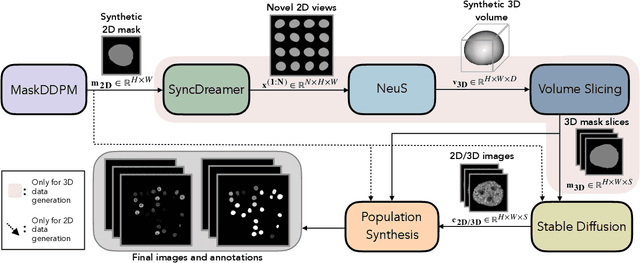
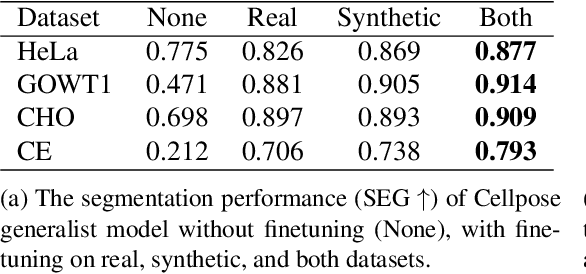
Abstract:Automated cell segmentation in microscopy images is essential for biomedical research, yet conventional methods are labor-intensive and prone to error. While deep learning-based approaches have proven effective, they often require large annotated datasets, which are scarce due to the challenges of manual annotation. To overcome this, we propose a novel framework for synthesizing densely annotated 2D and 3D cell microscopy images using cascaded diffusion models. Our method synthesizes 2D and 3D cell masks from sparse 2D annotations using multi-level diffusion models and NeuS, a 3D surface reconstruction approach. Following that, a pretrained 2D Stable Diffusion model is finetuned to generate realistic cell textures and the final outputs are combined to form cell populations. We show that training a segmentation model with a combination of our synthetic data and real data improves cell segmentation performance by up to 9\% across multiple datasets. Additionally, the FID scores indicate that the synthetic data closely resembles real data. The code for our proposed approach will be available at https://github.com/ruveydayilmaz0/cascaded_diffusion.
Visual Fixation-Based Retinal Prosthetic Simulation
Oct 15, 2024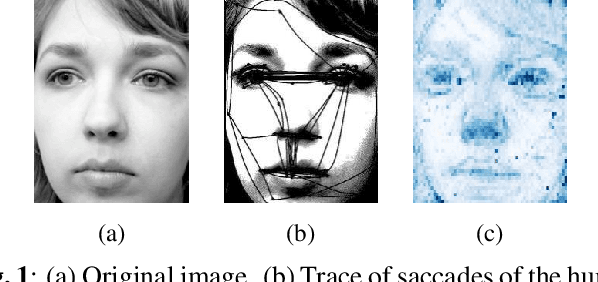
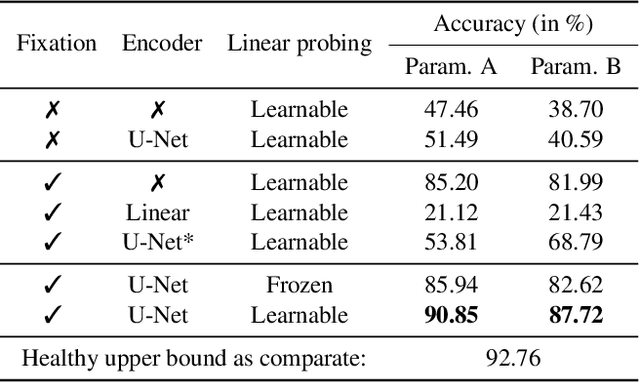
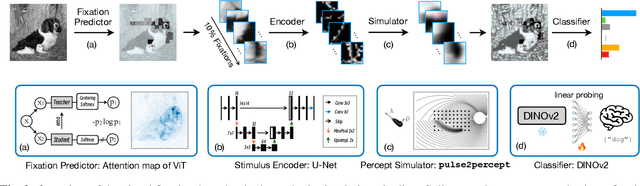
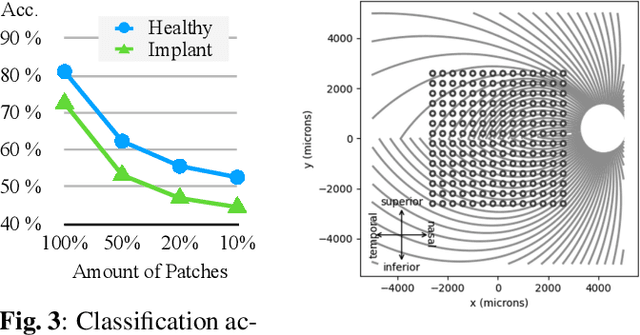
Abstract:This study proposes a retinal prosthetic simulation framework driven by visual fixations, inspired by the saccade mechanism, and assesses performance improvements through end-to-end optimization in a classification task. Salient patches are predicted from input images using the self-attention map of a vision transformer to mimic visual fixations. These patches are then encoded by a trainable U-Net and simulated using the pulse2percept framework to predict visual percepts. By incorporating a learnable encoder, we aim to optimize the visual information transmitted to the retinal implant, addressing both the limited resolution of the electrode array and the distortion between the input stimuli and resulting phosphenes. The predicted percepts are evaluated using the self-supervised DINOv2 foundation model, with an optional learnable linear layer for classification accuracy. On a subset of the ImageNet validation set, the fixation-based framework achieves a classification accuracy of 87.72%, using computational parameters based on a real subject's physiological data, significantly outperforming the downsampling-based accuracy of 40.59% and approaching the healthy upper bound of 92.76%. Our approach shows promising potential for producing more semantically understandable percepts with the limited resolution available in retinal prosthetics.
Annotated Biomedical Video Generation using Denoising Diffusion Probabilistic Models and Flow Fields
Mar 26, 2024



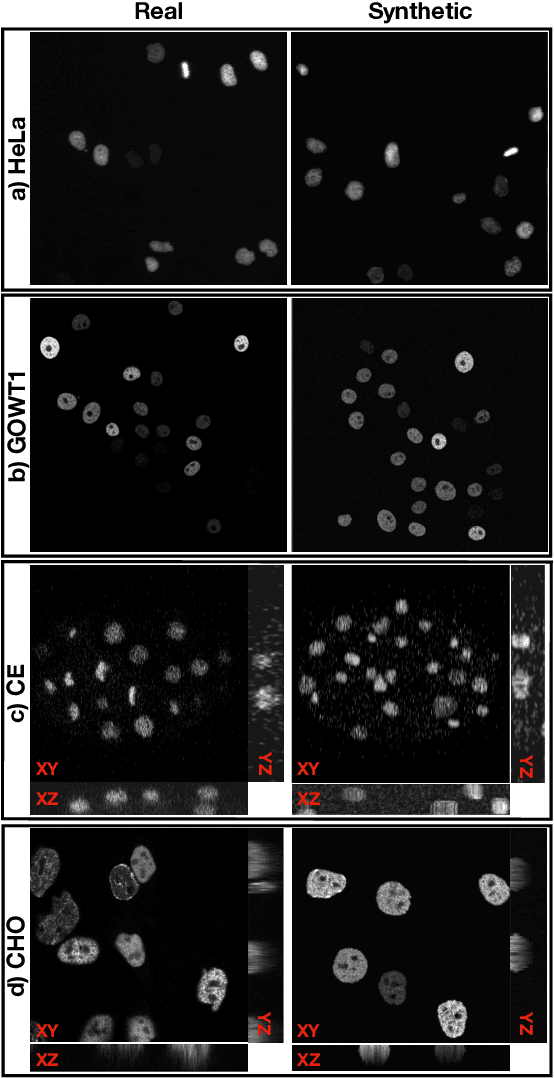
Abstract:The segmentation and tracking of living cells play a vital role within the biomedical domain, particularly in cancer research, drug development, and developmental biology. These are usually tedious and time-consuming tasks that are traditionally done by biomedical experts. Recently, to automatize these processes, deep learning based segmentation and tracking methods have been proposed. These methods require large-scale datasets and their full potential is constrained by the scarcity of annotated data in the biomedical imaging domain. To address this limitation, we propose Biomedical Video Diffusion Model (BVDM), capable of generating realistic-looking synthetic microscopy videos. Trained only on a single real video, BVDM can generate videos of arbitrary length with pixel-level annotations that can be used for training data-hungry models. It is composed of a denoising diffusion probabilistic model (DDPM) generating high-fidelity synthetic cell microscopy images and a flow prediction model (FPM) predicting the non-rigid transformation between consecutive video frames. During inference, initially, the DDPM imposes realistic cell textures on synthetic cell masks which are generated based on real data statistics. The flow prediction model predicts the flow field between consecutive masks and applies that to the DDPM output from the previous time frame to create the next one while keeping temporal consistency. BVDM outperforms state-of-the-art synthetic live cell microscopy video generation models. Furthermore, we demonstrate that a sufficiently large synthetic dataset enhances the performance of cell segmentation and tracking models compared to using a limited amount of available real data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge