Yoon Seong Choi
Evidence-empowered Transfer Learning for Alzheimer's Disease
Mar 03, 2023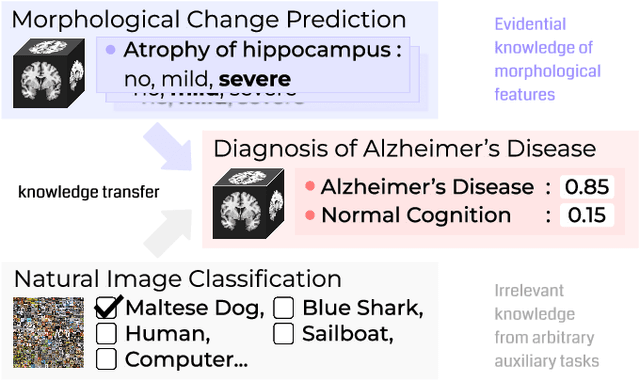
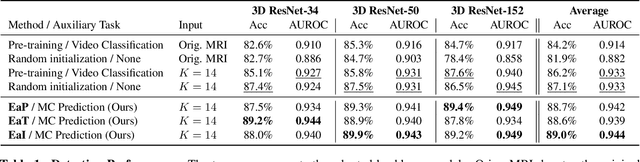
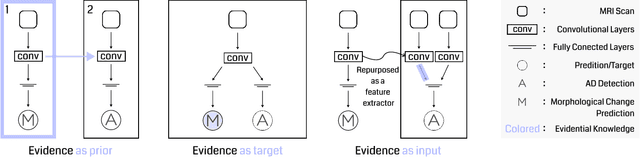
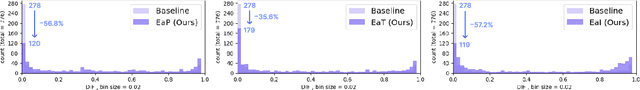
Abstract:Transfer learning has been widely utilized to mitigate the data scarcity problem in the field of Alzheimer's disease (AD). Conventional transfer learning relies on re-using models trained on AD-irrelevant tasks such as natural image classification. However, it often leads to negative transfer due to the discrepancy between the non-medical source and target medical domains. To address this, we present evidence-empowered transfer learning for AD diagnosis. Unlike conventional approaches, we leverage an AD-relevant auxiliary task, namely morphological change prediction, without requiring additional MRI data. In this auxiliary task, the diagnosis model learns the evidential and transferable knowledge from morphological features in MRI scans. Experimental results demonstrate that our framework is not only effective in improving detection performance regardless of model capacity, but also more data-efficient and faithful.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge