Yaozu Wu
A Call for Collaborative Intelligence: Why Human-Agent Systems Should Precede AI Autonomy
Jun 11, 2025Abstract:Recent improvements in large language models (LLMs) have led many researchers to focus on building fully autonomous AI agents. This position paper questions whether this approach is the right path forward, as these autonomous systems still have problems with reliability, transparency, and understanding the actual requirements of human. We suggest a different approach: LLM-based Human-Agent Systems (LLM-HAS), where AI works with humans rather than replacing them. By keeping human involved to provide guidance, answer questions, and maintain control, these systems can be more trustworthy and adaptable. Looking at examples from healthcare, finance, and software development, we show how human-AI teamwork can handle complex tasks better than AI working alone. We also discuss the challenges of building these collaborative systems and offer practical solutions. This paper argues that progress in AI should not be measured by how independent systems become, but by how well they can work with humans. The most promising future for AI is not in systems that take over human roles, but in those that enhance human capabilities through meaningful partnership.
A Survey on Large Language Model based Human-Agent Systems
May 01, 2025Abstract:Recent advances in large language models (LLMs) have sparked growing interest in building fully autonomous agents. However, fully autonomous LLM-based agents still face significant challenges, including limited reliability due to hallucinations, difficulty in handling complex tasks, and substantial safety and ethical risks, all of which limit their feasibility and trustworthiness in real-world applications. To overcome these limitations, LLM-based human-agent systems (LLM-HAS) incorporate human-provided information, feedback, or control into the agent system to enhance system performance, reliability and safety. This paper provides the first comprehensive and structured survey of LLM-HAS. It clarifies fundamental concepts, systematically presents core components shaping these systems, including environment & profiling, human feedback, interaction types, orchestration and communication, explores emerging applications, and discusses unique challenges and opportunities. By consolidating current knowledge and offering a structured overview, we aim to foster further research and innovation in this rapidly evolving interdisciplinary field. Paper lists and resources are available at https://github.com/HenryPengZou/Awesome-LLM-Based-Human-Agent-System-Papers.
Multi-Agent Autonomous Driving Systems with Large Language Models: A Survey of Recent Advances
Feb 24, 2025Abstract:Autonomous Driving Systems (ADSs) are revolutionizing transportation by reducing human intervention, improving operational efficiency, and enhancing safety. Large Language Models (LLMs), known for their exceptional planning and reasoning capabilities, have been integrated into ADSs to assist with driving decision-making. However, LLM-based single-agent ADSs face three major challenges: limited perception, insufficient collaboration, and high computational demands. To address these issues, recent advancements in LLM-based multi-agent ADSs have focused on improving inter-agent communication and cooperation. This paper provides a frontier survey of LLM-based multi-agent ADSs. We begin with a background introduction to related concepts, followed by a categorization of existing LLM-based approaches based on different agent interaction modes. We then discuss agent-human interactions in scenarios where LLM-based agents engage with humans. Finally, we summarize key applications, datasets, and challenges in this field to support future research (https://anonymous.4open.science/r/LLM-based_Multi-agent_ADS-3A5C/README.md).
Probing many-body Bell correlation depth with superconducting qubits
Jun 25, 2024


Abstract:Quantum nonlocality describes a stronger form of quantum correlation than that of entanglement. It refutes Einstein's belief of local realism and is among the most distinctive and enigmatic features of quantum mechanics. It is a crucial resource for achieving quantum advantages in a variety of practical applications, ranging from cryptography and certified random number generation via self-testing to machine learning. Nevertheless, the detection of nonlocality, especially in quantum many-body systems, is notoriously challenging. Here, we report an experimental certification of genuine multipartite Bell correlations, which signal nonlocality in quantum many-body systems, up to 24 qubits with a fully programmable superconducting quantum processor. In particular, we employ energy as a Bell correlation witness and variationally decrease the energy of a many-body system across a hierarchy of thresholds, below which an increasing Bell correlation depth can be certified from experimental data. As an illustrating example, we variationally prepare the low-energy state of a two-dimensional honeycomb model with 73 qubits and certify its Bell correlations by measuring an energy that surpasses the corresponding classical bound with up to 48 standard deviations. In addition, we variationally prepare a sequence of low-energy states and certify their genuine multipartite Bell correlations up to 24 qubits via energies measured efficiently by parity oscillation and multiple quantum coherence techniques. Our results establish a viable approach for preparing and certifying multipartite Bell correlations, which provide not only a finer benchmark beyond entanglement for quantum devices, but also a valuable guide towards exploiting multipartite Bell correlation in a wide spectrum of practical applications.
Experimental quantum adversarial learning with programmable superconducting qubits
Apr 04, 2022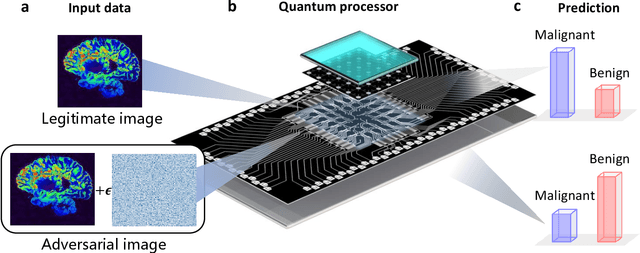
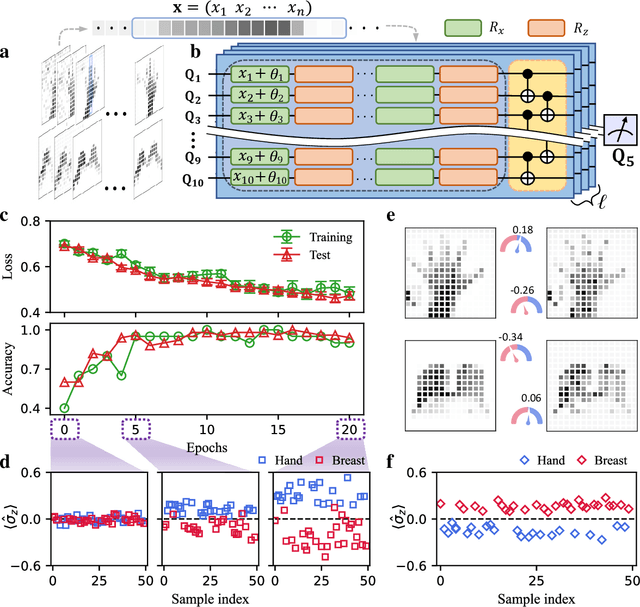
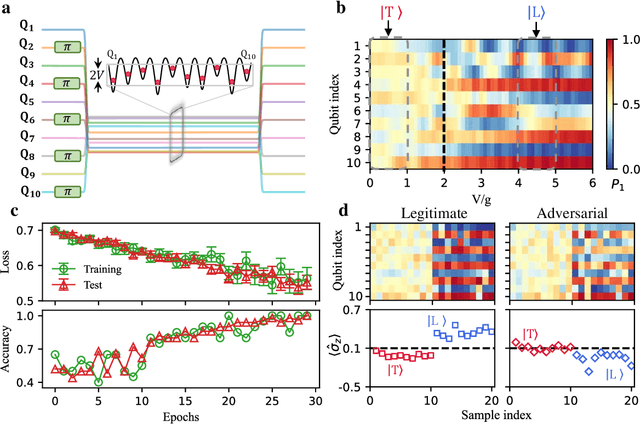
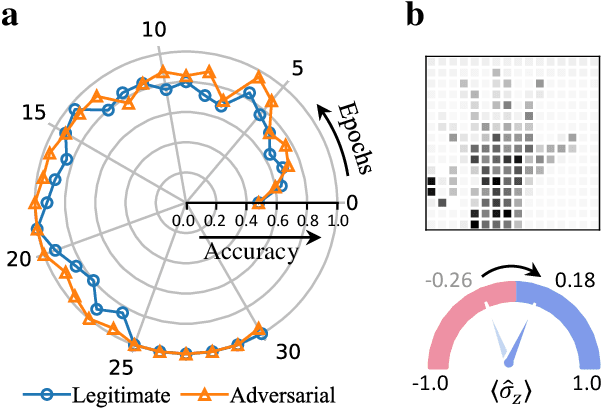
Abstract:Quantum computing promises to enhance machine learning and artificial intelligence. Different quantum algorithms have been proposed to improve a wide spectrum of machine learning tasks. Yet, recent theoretical works show that, similar to traditional classifiers based on deep classical neural networks, quantum classifiers would suffer from the vulnerability problem: adding tiny carefully-crafted perturbations to the legitimate original data samples would facilitate incorrect predictions at a notably high confidence level. This will pose serious problems for future quantum machine learning applications in safety and security-critical scenarios. Here, we report the first experimental demonstration of quantum adversarial learning with programmable superconducting qubits. We train quantum classifiers, which are built upon variational quantum circuits consisting of ten transmon qubits featuring average lifetimes of 150 $\mu$s, and average fidelities of simultaneous single- and two-qubit gates above 99.94% and 99.4% respectively, with both real-life images (e.g., medical magnetic resonance imaging scans) and quantum data. We demonstrate that these well-trained classifiers (with testing accuracy up to 99%) can be practically deceived by small adversarial perturbations, whereas an adversarial training process would significantly enhance their robustness to such perturbations. Our results reveal experimentally a crucial vulnerability aspect of quantum learning systems under adversarial scenarios and demonstrate an effective defense strategy against adversarial attacks, which provide a valuable guide for quantum artificial intelligence applications with both near-term and future quantum devices.
A Literature Review of Recent Graph Embedding Techniques for Biomedical Data
Jan 20, 2021


Abstract:With the rapid development of biomedical software and hardware, a large amount of relational data interlinking genes, proteins, chemical components, drugs, diseases, and symptoms has been collected for modern biomedical research. Many graph-based learning methods have been proposed to analyze such type of data, giving a deeper insight into the topology and knowledge behind the biomedical data, which greatly benefit to both academic research and industrial application for human healthcare. However, the main difficulty is how to handle high dimensionality and sparsity of the biomedical graphs. Recently, graph embedding methods provide an effective and efficient way to address the above issues. It converts graph-based data into a low dimensional vector space where the graph structural properties and knowledge information are well preserved. In this survey, we conduct a literature review of recent developments and trends in applying graph embedding methods for biomedical data. We also introduce important applications and tasks in the biomedical domain as well as associated public biomedical datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge