Yaohang Li
MMAC-Copilot: Multi-modal Agent Collaboration Operating System Copilot
Apr 28, 2024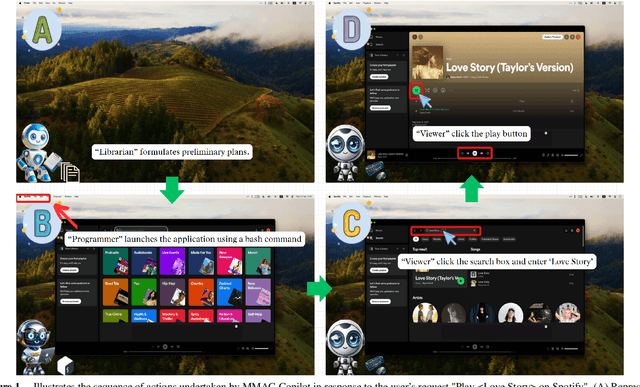
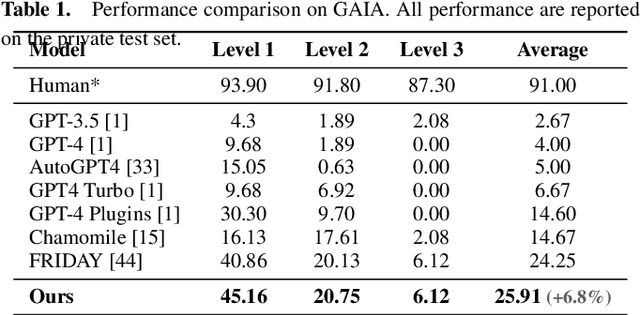
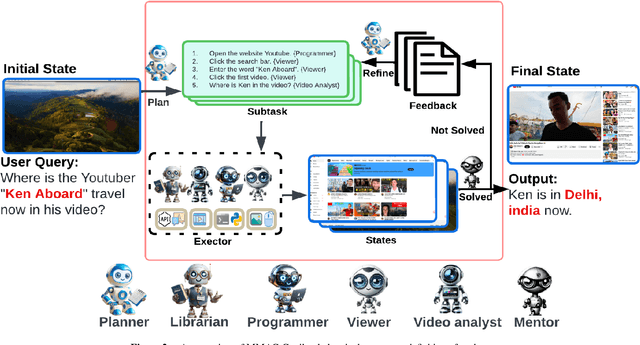

Abstract:Autonomous virtual agents are often limited by their singular mode of interaction with real-world environments, restricting their versatility. To address this, we propose the Multi-Modal Agent Collaboration framework (MMAC-Copilot), a framework utilizes the collective expertise of diverse agents to enhance interaction ability with operating systems. The framework introduces a team collaboration chain, enabling each participating agent to contribute insights based on their specific domain knowledge, effectively reducing the hallucination associated with knowledge domain gaps. To evaluate the performance of MMAC-Copilot, we conducted experiments using both the GAIA benchmark and our newly introduced Visual Interaction Benchmark (VIBench). VIBench focuses on non-API-interactable applications across various domains, including 3D gaming, recreation, and office scenarios. MMAC-Copilot achieved exceptional performance on GAIA, with an average improvement of 6.8\% over existing leading systems. Furthermore, it demonstrated remarkable capability on VIBench, particularly in managing various methods of interaction within systems and applications. These results underscore MMAC-Copilot's potential in advancing the field of autonomous virtual agents through its innovative approach to agent collaboration.
TripHLApan: predicting HLA molecules binding peptides based on triple coding matrix and transfer learning
Aug 06, 2022



Abstract:Human leukocyte antigen (HLA) is an important molecule family in the field of human immunity, which recognizes foreign threats and triggers immune responses by presenting peptides to T cells. In recent years, the synthesis of tumor vaccines to induce specific immune responses has become the forefront of cancer treatment. Computationally modeling the binding patterns between peptide and HLA can greatly accelerate the development of tumor vaccines. However, most of the prediction methods performance is very limited and they cannot fully take advantage of the analysis of existing biological knowledge as the basis of modeling. In this paper, we propose TripHLApan, a novel pan-specific prediction model, for HLA molecular peptide binding prediction. TripHLApan exhibits powerful prediction ability by integrating triple coding matrix, BiGRU + Attention models, and transfer learning strategy. The comprehensive evaluations demonstrate the effectiveness of TripHLApan in predicting HLA-I and HLA-II peptide binding in different test environments. The predictive power of HLA-I is further demonstrated in the latest data set. In addition, we show that TripHLApan has strong binding reconstitution ability in the samples of a melanoma patient. In conclusion, TripHLApan is a powerful tool for predicting the binding of HLA-I and HLA-II molecular peptides for the synthesis of tumor vaccines.
A survey of machine learning-based physics event generation
Jun 01, 2021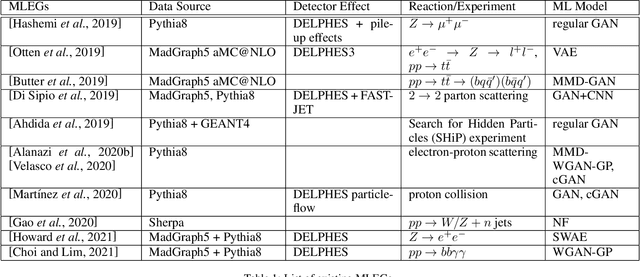
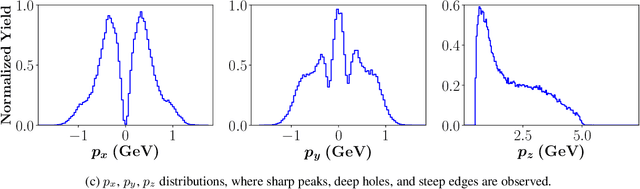
Abstract:Event generators in high-energy nuclear and particle physics play an important role in facilitating studies of particle reactions. We survey the state-of-the-art of machine learning (ML) efforts at building physics event generators. We review ML generative models used in ML-based event generators and their specific challenges, and discuss various approaches of incorporating physics into the ML model designs to overcome these challenges. Finally, we explore some open questions related to super-resolution, fidelity, and extrapolation for physics event generation based on ML technology.
Simulation of electron-proton scattering events by a Feature-Augmented and Transformed Generative Adversarial Network (FAT-GAN)
Jan 29, 2020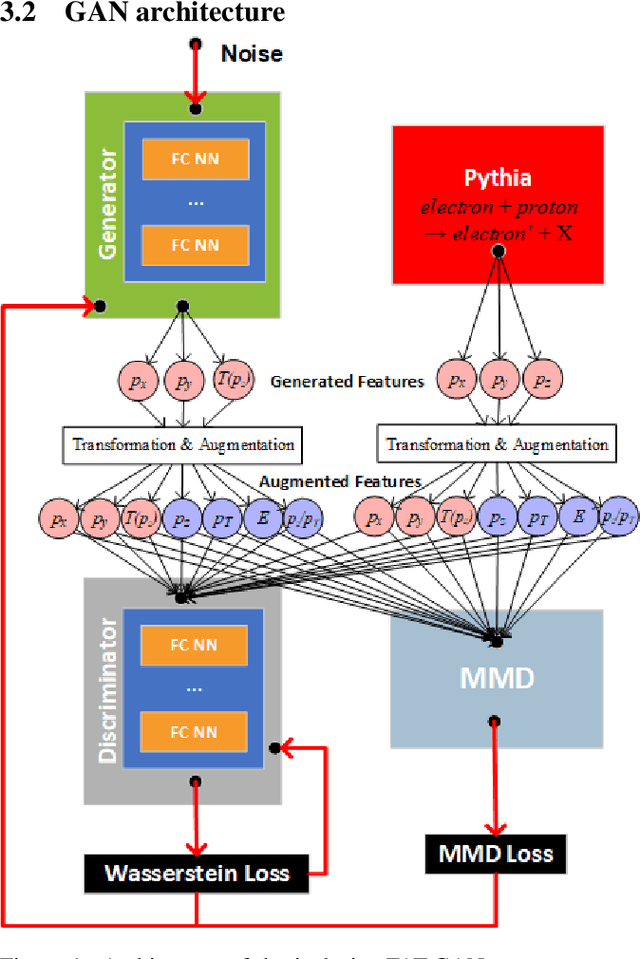
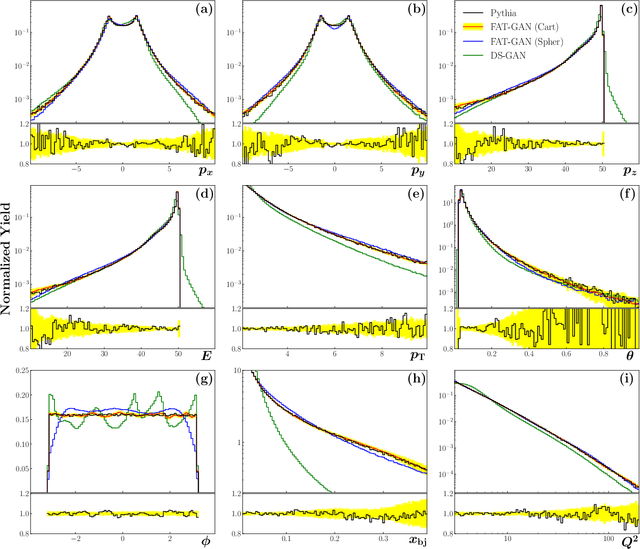
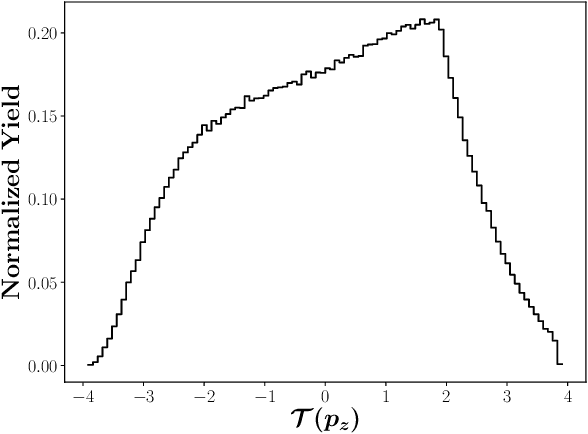
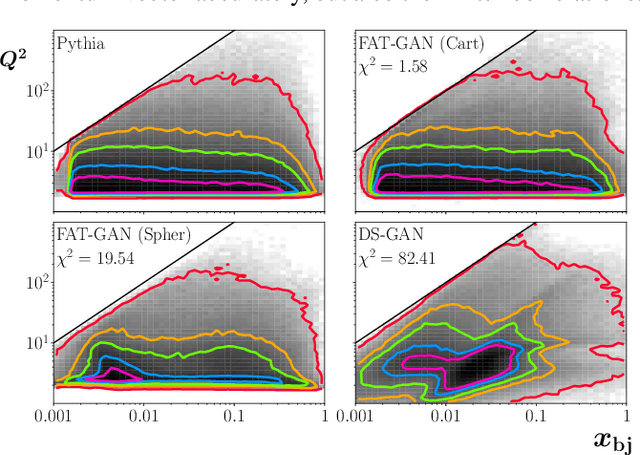
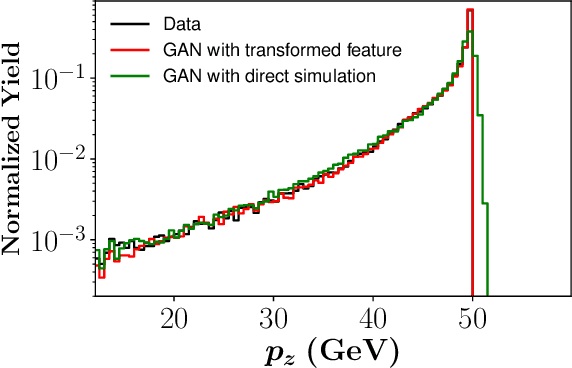
Abstract:We apply generative adversarial network (GAN) technology to build an event generator that simulates particle production in electron-proton scattering that is free of theoretical assumptions about underlying particle dynamics. The difficulty of efficiently training a GAN event simulator lies in learning the complicated patterns of the distributions of the particles physical properties. We develop a GAN that selects a set of transformed features from particle momenta that can be generated easily by the generator, and uses these to produce a set of augmented features that improve the sensitivity of the discriminator. The new Feature-Augmented and Transformed GAN (FAT-GAN) is able to faithfully reproduce the distribution of final state electron momenta in inclusive electron scattering, without the need for input derived from domain-based theoretical assumptions. The developed technology can play a significant role in boosting the science of the Jefferson Lab 12 GeV program and the future Electron-Ion Collider.
Faster Matrix Completion Using Randomized SVD
Oct 16, 2018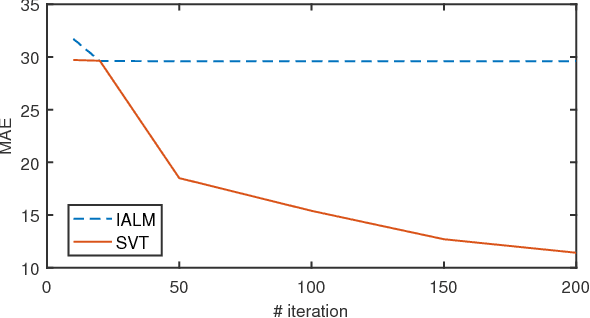
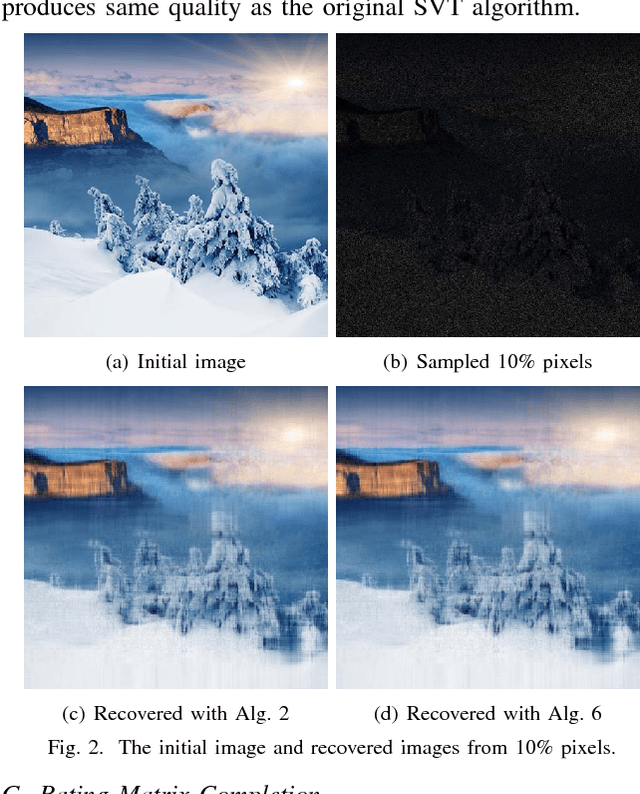
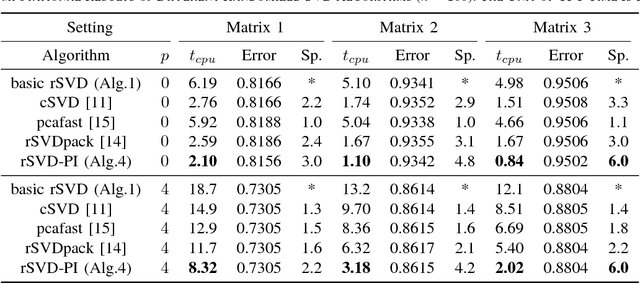
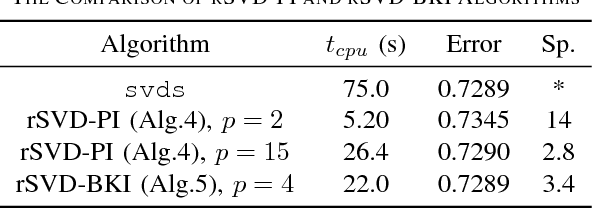
Abstract:Matrix completion is a widely used technique for image inpainting and personalized recommender system, etc. In this work, we focus on accelerating the matrix completion using faster randomized singular value decomposition (rSVD). Firstly, two fast randomized algorithms (rSVD-PI and rSVD- BKI) are proposed for handling sparse matrix. They make use of an eigSVD procedure and several accelerating skills. Then, with the rSVD-BKI algorithm and a new subspace recycling technique, we accelerate the singular value thresholding (SVT) method in [1] to realize faster matrix completion. Experiments show that the proposed rSVD algorithms can be 6X faster than the basic rSVD algorithm [2] while keeping same accuracy. For image inpainting and movie-rating estimation problems, the proposed accelerated SVT algorithm consumes 15X and 8X less CPU time than the methods using svds and lansvd respectively, without loss of accuracy.
Single-Pass PCA of Large High-Dimensional Data
Apr 25, 2017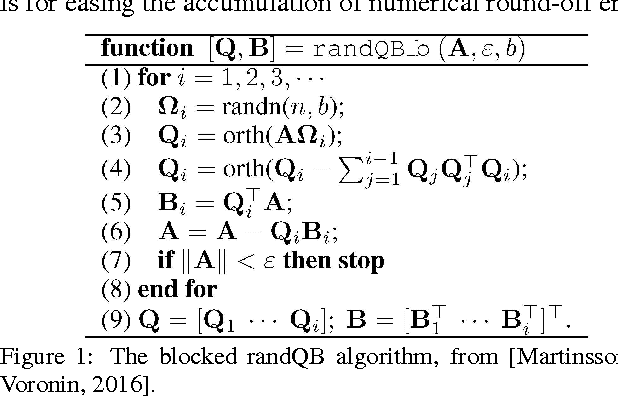
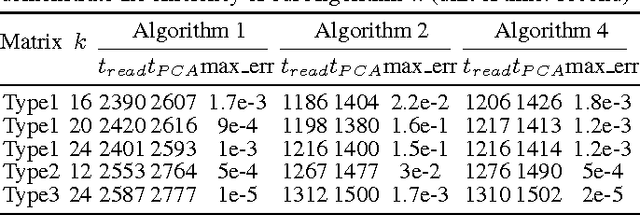
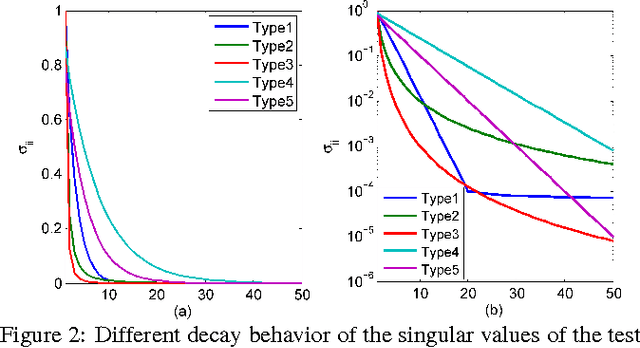
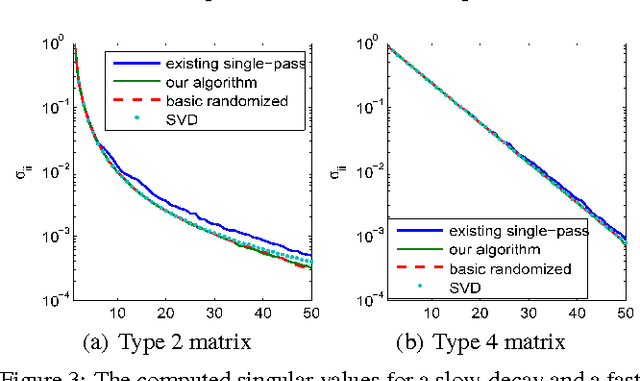
Abstract:Principal component analysis (PCA) is a fundamental dimension reduction tool in statistics and machine learning. For large and high-dimensional data, computing the PCA (i.e., the singular vectors corresponding to a number of dominant singular values of the data matrix) becomes a challenging task. In this work, a single-pass randomized algorithm is proposed to compute PCA with only one pass over the data. It is suitable for processing extremely large and high-dimensional data stored in slow memory (hard disk) or the data generated in a streaming fashion. Experiments with synthetic and real data validate the algorithm's accuracy, which has orders of magnitude smaller error than an existing single-pass algorithm. For a set of high-dimensional data stored as a 150 GB file, the proposed algorithm is able to compute the first 50 principal components in just 24 minutes on a typical 24-core computer, with less than 1 GB memory cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge