Yading Yuan
Federated prediction for scalable and privacy-preserved knowledge-based planning in radiotherapy
May 20, 2025Abstract:Background: Deep learning has potential to improve the efficiency and consistency of radiation therapy planning, but clinical adoption is hindered by the limited model generalizability due to data scarcity and heterogeneity among institutions. Although aggregating data from different institutions could alleviate this problem, data sharing is a practical challenge due to concerns about patient data privacy and other technical obstacles. Purpose: This work aims to address this dilemma by developing FedKBP+, a comprehensive federated learning (FL) platform for predictive tasks in real-world applications in radiotherapy treatment planning. Methods: We implemented a unified communication stack based on Google Remote Procedure Call (gRPC) to support communication between participants whether located on the same workstation or distributed across multiple workstations. In addition to supporting the centralized FL strategies commonly available in existing open-source frameworks, FedKBP+ also provides a fully decentralized FL model where participants directly exchange model weights to each other through Peer-to-Peer communication. We evaluated FedKBP+ on three predictive tasks using scale-attention network (SA-Net) as the predictive model. Conclusions: Our results demonstrate that FedKBP+ is highly effective, efficient and robust, showing great potential as a federated learning platform for radiation therapy.
CAST: Time-Varying Treatment Effects with Application to Chemotherapy and Radiotherapy on Head and Neck Squamous Cell Carcinoma
May 09, 2025Abstract:Causal machine learning (CML) enables individualized estimation of treatment effects, offering critical advantages over traditional correlation-based methods. However, existing approaches for medical survival data with censoring such as causal survival forests estimate effects at fixed time points, limiting their ability to capture dynamic changes over time. We introduce Causal Analysis for Survival Trajectories (CAST), a novel framework that models treatment effects as continuous functions of time following treatment. By combining parametric and non-parametric methods, CAST overcomes the limitations of discrete time-point analysis to estimate continuous effect trajectories. Using the RADCURE dataset [1] of 2,651 patients with head and neck squamous cell carcinoma (HNSCC) as a clinically relevant example, CAST models how chemotherapy and radiotherapy effects evolve over time at the population and individual levels. By capturing the temporal dynamics of treatment response, CAST reveals how treatment effects rise, peak, and decline over the follow-up period, helping clinicians determine when and for whom treatment benefits are maximized. This framework advances the application of CML to personalized care in HNSCC and other life-threatening medical conditions. Source code/data available at: https://github.com/CAST-FW/HNSCC
FedKBP: Federated dose prediction framework for knowledge-based planning in radiation therapy
Aug 17, 2024

Abstract:Dose prediction plays a key role in knowledge-based planning (KBP) by automatically generating patient-specific dose distribution. Recent advances in deep learning-based dose prediction methods necessitates collaboration among data contributors for improved performance. Federated learning (FL) has emerged as a solution, enabling medical centers to jointly train deep-learning models without compromising patient data privacy. We developed the FedKBP framework to evaluate the performances of centralized, federated, and individual (i.e. separated) training of dose prediction model on the 340 plans from OpenKBP dataset. To simulate FL and individual training, we divided the data into 8 training sites. To evaluate the effect of inter-site data variation on model training, we implemented two types of case distributions: 1) Independent and identically distributed (IID), where the training and validating cases were evenly divided among the 8 sites, and 2) non-IID, where some sites have more cases than others. The results show FL consistently outperforms individual training on both model optimization speed and out-of-sample testing scores, highlighting the advantage of FL over individual training. Under IID data division, FL shows comparable performance to centralized training, underscoring FL as a promising alternative to traditional pooled-data training. Under non-IID division, larger sites outperformed smaller sites by up to 19% on testing scores, confirming the need of collaboration among data owners to achieve better prediction accuracy. Meanwhile, non-IID FL showed reduced performance as compared to IID FL, posing the need for more sophisticated FL method beyond mere model averaging to handle data variation among participating sites.
Decentralized Gossip Mutual Learning (GML) for brain tumor segmentation on multi-parametric MRI
Jan 27, 2024

Abstract:Federated Learning (FL) enables collaborative model training among medical centers without sharing private data. However, traditional FL risks on server failures and suboptimal performance on local data due to the nature of centralized model aggregation. To address these issues, we present Gossip Mutual Learning (GML), a decentralized framework that uses Gossip Protocol for direct peer-to-peer communication. In addition, GML encourages each site to optimize its local model through mutual learning to account for data variations among different sites. For the task of tumor segmentation using 146 cases from four clinical sites in BraTS 2021 dataset, we demonstrated GML outperformed local models and achieved similar performance as FedAvg with only 25% communication overhead.
Decentralized Gossip Mutual Learning for automatic head and neck tumor segmentation
Jan 11, 2024Abstract:Federated learning (FL) has emerged as a promising strategy for collaboratively training complicated machine learning models from different medical centers without the need of data sharing. However, the traditional FL relies on a central server to orchestrate the global model training among clients. This makes it vulnerable to the failure of the model server. Meanwhile, the model trained based on the global data property may not yield the best performance on the local data of a particular site due to the variations of data characteristics among them. To address these limitations, we proposed Gossip Mutual Learning(GML), a decentralized collaborative learning framework that employs Gossip Protocol for direct peer-to-peer communication and encourages each site to optimize its local model by leveraging useful information from peers through mutual learning. On the task of tumor segmentation on PET/CT images using HECKTOR21 dataset with 223 cases from five clinical sites, we demonstrated GML could improve tumor segmentation performance in terms of Dice Similarity Coefficient (DSC) by 3.2%, 4.6% and 10.4% on site-specific testing cases as compared to three baseline methods: pooled training, FedAvg and individual training, respectively. We also showed GML has comparable generalization performance as pooled training and FedAvg when applying them on 78 cases from two out-of-sample sites where no case was used for model training. In our experimental setup, GML showcased a sixfold decrease in communication overhead compared to FedAvg, requiring only 16.67% of the total communication overhead.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
Automatic Brain Tumor Segmentation with Scale Attention Network
Nov 10, 2020



Abstract:Automatic segmentation of brain tumors is an essential but challenging step for extracting quantitative imaging biomarkers for accurate tumor detection, diagnosis, prognosis, treatment planning and assessment. Multimodal Brain Tumor Segmentation Challenge 2020 (BraTS 2020) provides a common platform for comparing different automatic algorithms on multi-parametric Magnetic Resonance Imaging (mpMRI) in tasks of 1) Brain tumor segmentation MRI scans; 2) Prediction of patient overall survival (OS) from pre-operative MRI scans; 3) Distinction of true tumor recurrence from treatment related effects and 4) Evaluation of uncertainty measures in segmentation. We participate the image segmentation challenge by developing a fully automatic segmentation network based on encoder-decoder architecture. In order to better integrate information across different scales, we propose a dynamic scale attention mechanism that incorporates low-level details with high-level semantics from feature maps at different scales. Our framework was trained using the 369 challenge training cases provided by BraTS 2020, and achieved an average Dice Similarity Coefficient (DSC) of 0.8828, 0.8433 and 0.8177, as well as 95% Hausdorff distance (in millimeter) of 5.2176, 17.9697 and 13.4298 on 166 testing cases for whole tumor, tumor core and enhanced tumor, respectively, which ranked itself as the 3rd place in the Brats 2020 challenge.
A Deep Regression Model for Seed Identification in Prostate Brachytherapy
Jun 24, 2019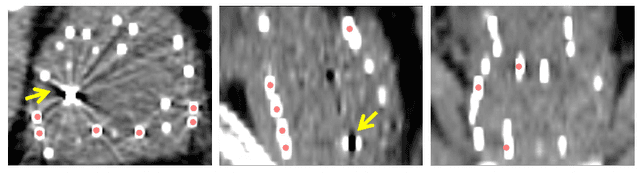
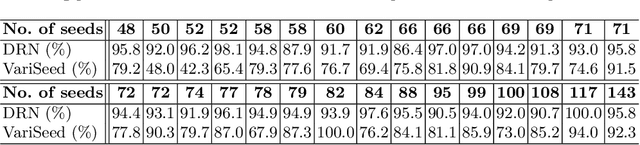
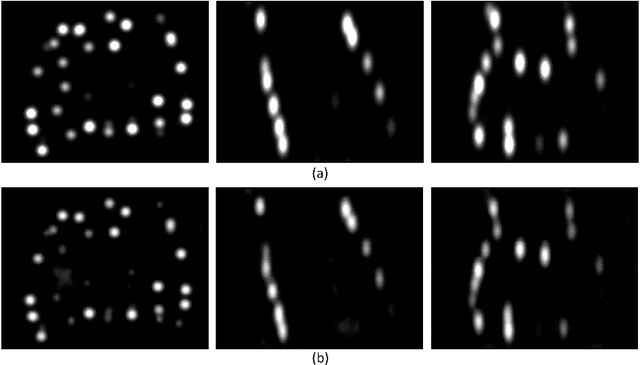
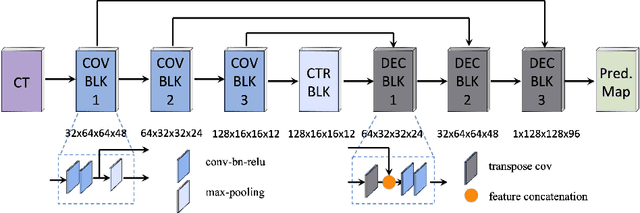
Abstract:Post-implant dosimetry (PID) is an essential step of prostate brachytherapy that utilizes CT to image the prostate and allow the location and dose distribution of the radioactive seeds to be directly related to the actual prostate. However, it it a very challenging task to identify these seeds in CT images due to the severe metal artifacts and high-overlapped appearance when multiple seeds clustered together. In this paper, we propose an automatic and efficient algorithm based on 3D deep fully convolutional network for identifying implanted seeds in CT images. Our method models the seed localization task as a supervised regression problem that projects the input CT image to a map where each element represents the probability that the corresponding input voxel belongs to a seed. This deep regression model significantly suppresses image artifacts and makes the post-processing much easier and more controllable. The proposed method is validated on a large clinical database with 7820 seeds in 100 patients, in which 5534 seeds from 70 patients were used for model training and validation. Our method correctly detected 2150 of 2286 (94.1%) seeds in the 30 testing patients, yielding 16% improvement as compared to a widely-used commercial seed finder software (VariSeed, Varian, Palo Alto, CA).
Hierarchical Convolutional-Deconvolutional Neural Networks for Automatic Liver and Tumor Segmentation
Oct 12, 2017Abstract:Automatic segmentation of liver and its tumors is an essential step for extracting quantitative imaging biomarkers for accurate tumor detection, diagnosis, prognosis and assessment of tumor response to treatment. MICCAI 2017 Liver Tumor Segmentation Challenge (LiTS) provides a common platform for comparing different automatic algorithms on contrast-enhanced abdominal CT images in tasks including 1) liver segmentation, 2) liver tumor segmentation, and 3) tumor burden estimation. We participate this challenge by developing a hierarchical framework based on deep fully convolutional-deconvolutional neural networks (CDNN). A simple CDNN model is firstly trained to provide a quick but coarse segmentation of the liver on the entire CT volume, then another CDNN is applied to the liver region for fine liver segmentation. At last, the segmented liver region, which is enhanced by histogram equalization, is employed as an additional input to the third CDNN for tumor segmentation. Jaccard distance is used as loss function when training CDNN models to eliminate the need of sample re-weighting. Our framework is trained using the 130 challenge training cases provided by LiTS. The evaluation on the 70 challenge testing cases resulted in a mean Dice Similarity Coefficient (DSC) of 0.963 for liver segmentation, a mean DSC of 0.657 for tumor segmentation, and a root mean square error (RMSE) of 0.017 for tumor burden estimation, which ranked our method in the first, fifth and third place, respectively
Automatic skin lesion segmentation with fully convolutional-deconvolutional networks
Sep 28, 2017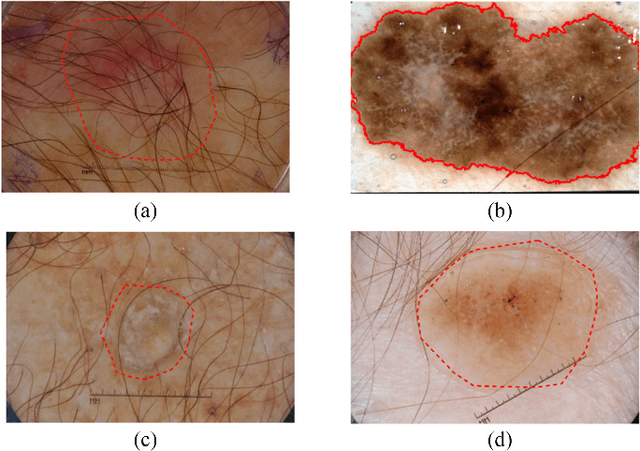
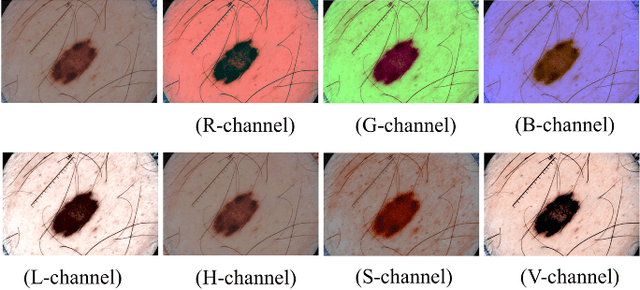
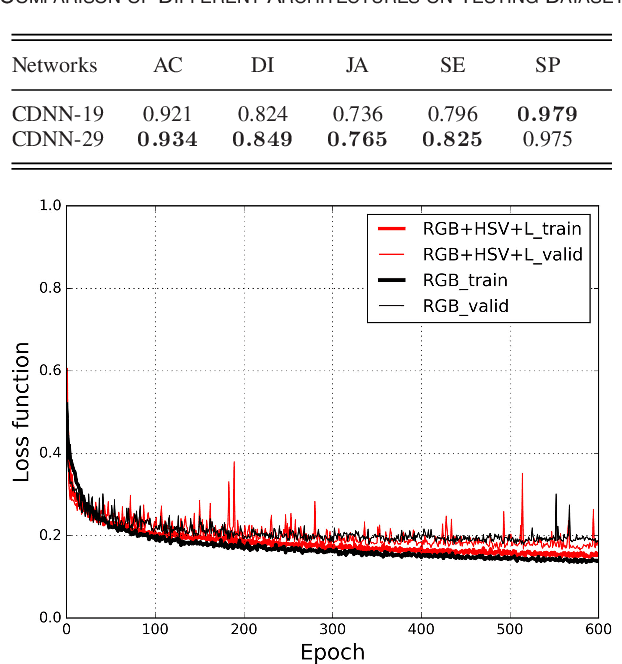
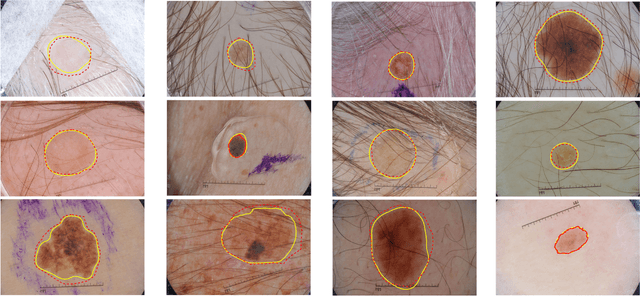
Abstract:This paper summarizes our method and validation results for the ISBI Challenge 2017 - Skin Lesion Analysis Towards Melanoma Detection - Part I: Lesion Segmentation
* ISIC2017 challenge, 4 pages
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge