Jingyun Chen
Federated prediction for scalable and privacy-preserved knowledge-based planning in radiotherapy
May 20, 2025Abstract:Background: Deep learning has potential to improve the efficiency and consistency of radiation therapy planning, but clinical adoption is hindered by the limited model generalizability due to data scarcity and heterogeneity among institutions. Although aggregating data from different institutions could alleviate this problem, data sharing is a practical challenge due to concerns about patient data privacy and other technical obstacles. Purpose: This work aims to address this dilemma by developing FedKBP+, a comprehensive federated learning (FL) platform for predictive tasks in real-world applications in radiotherapy treatment planning. Methods: We implemented a unified communication stack based on Google Remote Procedure Call (gRPC) to support communication between participants whether located on the same workstation or distributed across multiple workstations. In addition to supporting the centralized FL strategies commonly available in existing open-source frameworks, FedKBP+ also provides a fully decentralized FL model where participants directly exchange model weights to each other through Peer-to-Peer communication. We evaluated FedKBP+ on three predictive tasks using scale-attention network (SA-Net) as the predictive model. Conclusions: Our results demonstrate that FedKBP+ is highly effective, efficient and robust, showing great potential as a federated learning platform for radiation therapy.
FedKBP: Federated dose prediction framework for knowledge-based planning in radiation therapy
Aug 17, 2024

Abstract:Dose prediction plays a key role in knowledge-based planning (KBP) by automatically generating patient-specific dose distribution. Recent advances in deep learning-based dose prediction methods necessitates collaboration among data contributors for improved performance. Federated learning (FL) has emerged as a solution, enabling medical centers to jointly train deep-learning models without compromising patient data privacy. We developed the FedKBP framework to evaluate the performances of centralized, federated, and individual (i.e. separated) training of dose prediction model on the 340 plans from OpenKBP dataset. To simulate FL and individual training, we divided the data into 8 training sites. To evaluate the effect of inter-site data variation on model training, we implemented two types of case distributions: 1) Independent and identically distributed (IID), where the training and validating cases were evenly divided among the 8 sites, and 2) non-IID, where some sites have more cases than others. The results show FL consistently outperforms individual training on both model optimization speed and out-of-sample testing scores, highlighting the advantage of FL over individual training. Under IID data division, FL shows comparable performance to centralized training, underscoring FL as a promising alternative to traditional pooled-data training. Under non-IID division, larger sites outperformed smaller sites by up to 19% on testing scores, confirming the need of collaboration among data owners to achieve better prediction accuracy. Meanwhile, non-IID FL showed reduced performance as compared to IID FL, posing the need for more sophisticated FL method beyond mere model averaging to handle data variation among participating sites.
H2ASeg: Hierarchical Adaptive Interaction and Weighting Network for Tumor Segmentation in PET/CT Images
Mar 28, 2024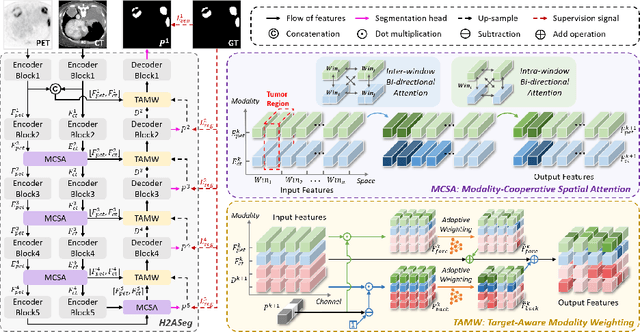
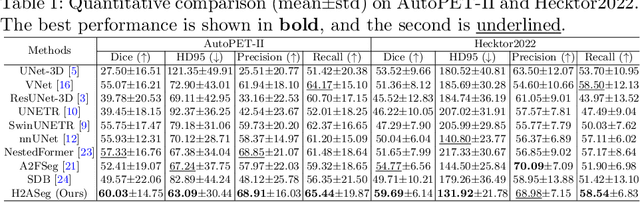
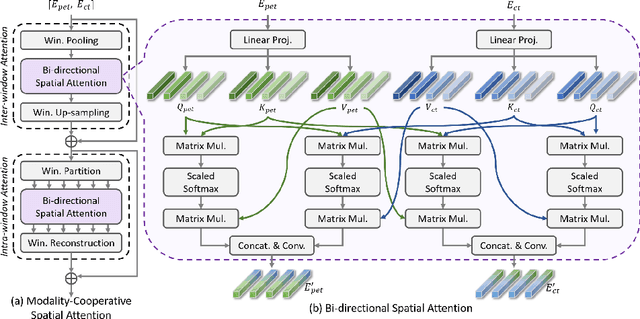
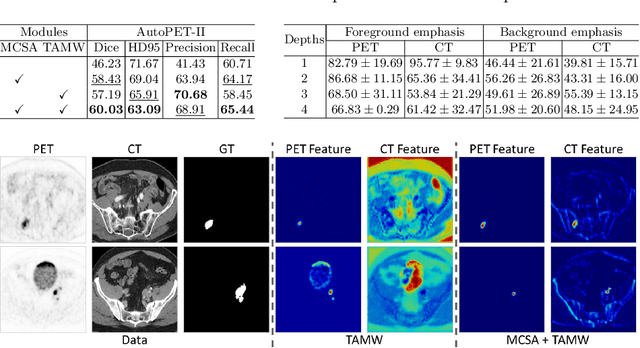
Abstract:Positron emission tomography (PET) combined with computed tomography (CT) imaging is routinely used in cancer diagnosis and prognosis by providing complementary information. Automatically segmenting tumors in PET/CT images can significantly improve examination efficiency. Traditional multi-modal segmentation solutions mainly rely on concatenation operations for modality fusion, which fail to effectively model the non-linear dependencies between PET and CT modalities. Recent studies have investigated various approaches to optimize the fusion of modality-specific features for enhancing joint representations. However, modality-specific encoders used in these methods operate independently, inadequately leveraging the synergistic relationships inherent in PET and CT modalities, for example, the complementarity between semantics and structure. To address these issues, we propose a Hierarchical Adaptive Interaction and Weighting Network termed H2ASeg to explore the intrinsic cross-modal correlations and transfer potential complementary information. Specifically, we design a Modality-Cooperative Spatial Attention (MCSA) module that performs intra- and inter-modal interactions globally and locally. Additionally, a Target-Aware Modality Weighting (TAMW) module is developed to highlight tumor-related features within multi-modal features, thereby refining tumor segmentation. By embedding these modules across different layers, H2ASeg can hierarchically model cross-modal correlations, enabling a nuanced understanding of both semantic and structural tumor features. Extensive experiments demonstrate the superiority of H2ASeg, outperforming state-of-the-art methods on AutoPet-II and Hecktor2022 benchmarks. The code is released at https://github.com/JinPLu/H2ASeg.
Decentralized Gossip Mutual Learning (GML) for brain tumor segmentation on multi-parametric MRI
Jan 27, 2024

Abstract:Federated Learning (FL) enables collaborative model training among medical centers without sharing private data. However, traditional FL risks on server failures and suboptimal performance on local data due to the nature of centralized model aggregation. To address these issues, we present Gossip Mutual Learning (GML), a decentralized framework that uses Gossip Protocol for direct peer-to-peer communication. In addition, GML encourages each site to optimize its local model through mutual learning to account for data variations among different sites. For the task of tumor segmentation using 146 cases from four clinical sites in BraTS 2021 dataset, we demonstrated GML outperformed local models and achieved similar performance as FedAvg with only 25% communication overhead.
Decentralized Gossip Mutual Learning for automatic head and neck tumor segmentation
Jan 11, 2024Abstract:Federated learning (FL) has emerged as a promising strategy for collaboratively training complicated machine learning models from different medical centers without the need of data sharing. However, the traditional FL relies on a central server to orchestrate the global model training among clients. This makes it vulnerable to the failure of the model server. Meanwhile, the model trained based on the global data property may not yield the best performance on the local data of a particular site due to the variations of data characteristics among them. To address these limitations, we proposed Gossip Mutual Learning(GML), a decentralized collaborative learning framework that employs Gossip Protocol for direct peer-to-peer communication and encourages each site to optimize its local model by leveraging useful information from peers through mutual learning. On the task of tumor segmentation on PET/CT images using HECKTOR21 dataset with 223 cases from five clinical sites, we demonstrated GML could improve tumor segmentation performance in terms of Dice Similarity Coefficient (DSC) by 3.2%, 4.6% and 10.4% on site-specific testing cases as compared to three baseline methods: pooled training, FedAvg and individual training, respectively. We also showed GML has comparable generalization performance as pooled training and FedAvg when applying them on 78 cases from two out-of-sample sites where no case was used for model training. In our experimental setup, GML showcased a sixfold decrease in communication overhead compared to FedAvg, requiring only 16.67% of the total communication overhead.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge