Songhan Jiang
PathReasoner-R1: Instilling Structured Reasoning into Pathology Vision-Language Model via Knowledge-Guided Policy Optimization
Jan 29, 2026Abstract:Vision-Language Models (VLMs) are advancing computational pathology with superior visual understanding capabilities. However, current systems often reduce diagnosis to directly output conclusions without verifiable evidence-linked reasoning, which severely limits clinical trust and hinders expert error rectification. To address these barriers, we construct PathReasoner, the first large-scale dataset of whole-slide image (WSI) reasoning. Unlike previous work reliant on unverified distillation, we develop a rigorous knowledge-guided generation pipeline. By leveraging medical knowledge graphs, we explicitly align structured pathological findings and clinical reasoning with diagnoses, generating over 20K high-quality instructional samples. Based on the database, we propose PathReasoner-R1, which synergizes trajectory-masked supervised fine-tuning with reasoning-oriented reinforcement learning to instill structured chain-of-thought capabilities. To ensure medical rigor, we engineer a knowledge-aware multi-granular reward function incorporating an Entity Reward mechanism strictly aligned with knowledge graphs. This effectively guides the model to optimize for logical consistency rather than mere outcome matching, thereby enhancing robustness. Extensive experiments demonstrate that PathReasoner-R1 achieves state-of-the-art performance on both PathReasoner and public benchmarks across various image scales, equipping pathology models with transparent, clinically grounded reasoning capabilities. Dataset and code are available at https://github.com/cyclexfy/PathReasoner-R1.
PathFLIP: Fine-grained Language-Image Pretraining for Versatile Computational Pathology
Dec 19, 2025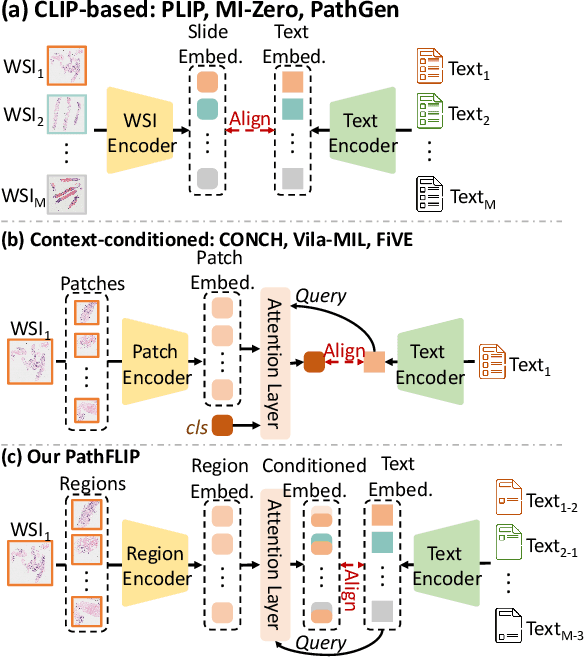
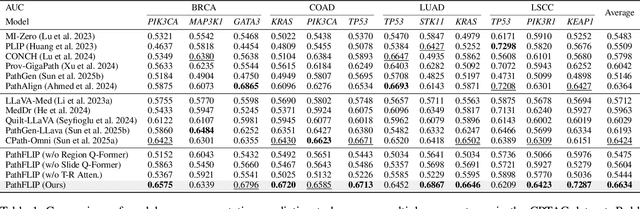
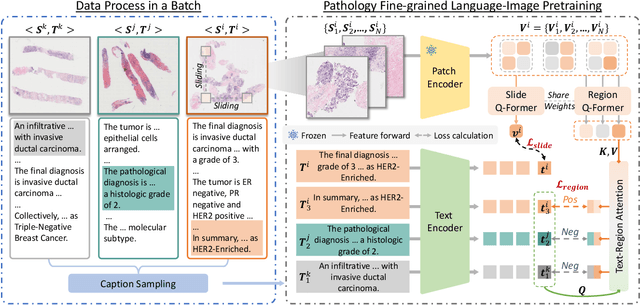
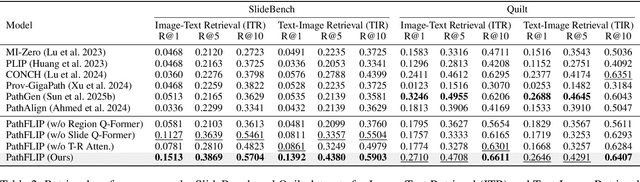
Abstract:While Vision-Language Models (VLMs) have achieved notable progress in computational pathology (CPath), the gigapixel scale and spatial heterogeneity of Whole Slide Images (WSIs) continue to pose challenges for multimodal understanding. Existing alignment methods struggle to capture fine-grained correspondences between textual descriptions and visual cues across thousands of patches from a slide, compromising their performance on downstream tasks. In this paper, we propose PathFLIP (Pathology Fine-grained Language-Image Pretraining), a novel framework for holistic WSI interpretation. PathFLIP decomposes slide-level captions into region-level subcaptions and generates text-conditioned region embeddings to facilitate precise visual-language grounding. By harnessing Large Language Models (LLMs), PathFLIP can seamlessly follow diverse clinical instructions and adapt to varied diagnostic contexts. Furthermore, it exhibits versatile capabilities across multiple paradigms, efficiently handling slide-level classification and retrieval, fine-grained lesion localization, and instruction following. Extensive experiments demonstrate that PathFLIP outperforms existing large-scale pathological VLMs on four representative benchmarks while requiring significantly less training data, paving the way for fine-grained, instruction-aware WSI interpretation in clinical practice.
Johnson-Lindenstrauss Lemma Guided Network for Efficient 3D Medical Segmentation
Sep 26, 2025Abstract:Lightweight 3D medical image segmentation remains constrained by a fundamental "efficiency / robustness conflict", particularly when processing complex anatomical structures and heterogeneous modalities. In this paper, we study how to redesign the framework based on the characteristics of high-dimensional 3D images, and explore data synergy to overcome the fragile representation of lightweight methods. Our approach, VeloxSeg, begins with a deployable and extensible dual-stream CNN-Transformer architecture composed of Paired Window Attention (PWA) and Johnson-Lindenstrauss lemma-guided convolution (JLC). For each 3D image, we invoke a "glance-and-focus" principle, where PWA rapidly retrieves multi-scale information, and JLC ensures robust local feature extraction with minimal parameters, significantly enhancing the model's ability to operate with low computational budget. Followed by an extension of the dual-stream architecture that incorporates modal interaction into the multi-scale image-retrieval process, VeloxSeg efficiently models heterogeneous modalities. Finally, Spatially Decoupled Knowledge Transfer (SDKT) via Gram matrices injects the texture prior extracted by a self-supervised network into the segmentation network, yielding stronger representations than baselines at no extra inference cost. Experimental results on multimodal benchmarks show that VeloxSeg achieves a 26% Dice improvement, alongside increasing GPU throughput by 11x and CPU by 48x. Codes are available at https://github.com/JinPLu/VeloxSeg.
IPGPhormer: Interpretable Pathology Graph-Transformer for Survival Analysis
Aug 17, 2025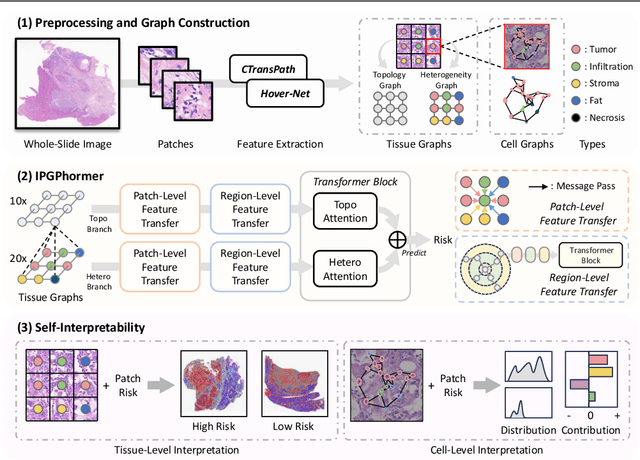
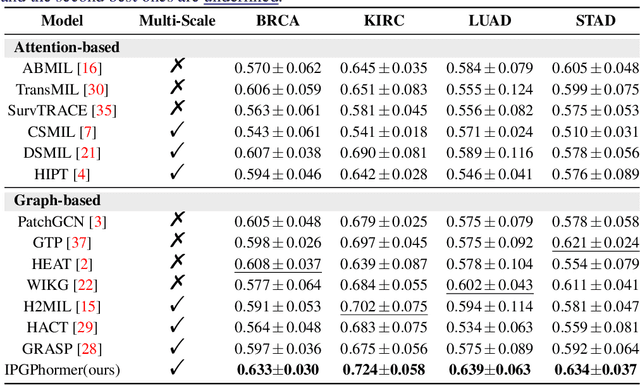
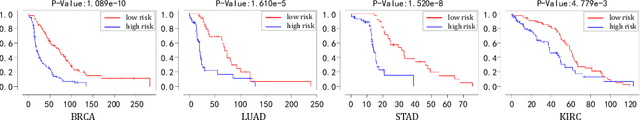
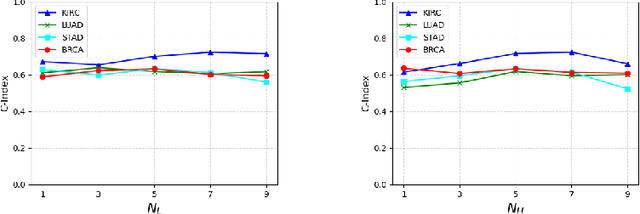
Abstract:Pathological images play an essential role in cancer prognosis, while survival analysis, which integrates computational techniques, can predict critical clinical events such as patient mortality or disease recurrence from whole-slide images (WSIs). Recent advancements in multiple instance learning have significantly improved the efficiency of survival analysis. However, existing methods often struggle to balance the modeling of long-range spatial relationships with local contextual dependencies and typically lack inherent interpretability, limiting their clinical utility. To address these challenges, we propose the Interpretable Pathology Graph-Transformer (IPGPhormer), a novel framework that captures the characteristics of the tumor microenvironment and models their spatial dependencies across the tissue. IPGPhormer uniquely provides interpretability at both tissue and cellular levels without requiring post-hoc manual annotations, enabling detailed analyses of individual WSIs and cross-cohort assessments. Comprehensive evaluations on four public benchmark datasets demonstrate that IPGPhormer outperforms state-of-the-art methods in both predictive accuracy and interpretability. In summary, our method, IPGPhormer, offers a promising tool for cancer prognosis assessment, paving the way for more reliable and interpretable decision-support systems in pathology. The code is publicly available at https://anonymous.4open.science/r/IPGPhormer-6EEB.
Multimodal Cross-Task Interaction for Survival Analysis in Whole Slide Pathological Images
Jun 25, 2024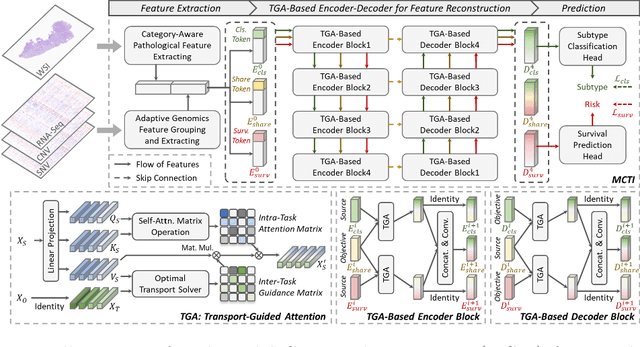
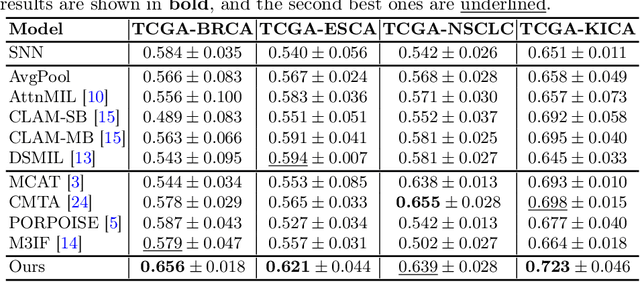

Abstract:Survival prediction, utilizing pathological images and genomic profiles, is increasingly important in cancer analysis and prognosis. Despite significant progress, precise survival analysis still faces two main challenges: (1) The massive pixels contained in whole slide images (WSIs) complicate the process of pathological images, making it difficult to generate an effective representation of the tumor microenvironment (TME). (2) Existing multimodal methods often rely on alignment strategies to integrate complementary information, which may lead to information loss due to the inherent heterogeneity between pathology and genes. In this paper, we propose a Multimodal Cross-Task Interaction (MCTI) framework to explore the intrinsic correlations between subtype classification and survival analysis tasks. Specifically, to capture TME-related features in WSIs, we leverage the subtype classification task to mine tumor regions. Simultaneously, multi-head attention mechanisms are applied in genomic feature extraction, adaptively performing genes grouping to obtain task-related genomic embedding. With the joint representation of pathological images and genomic data, we further introduce a Transport-Guided Attention (TGA) module that uses optimal transport theory to model the correlation between subtype classification and survival analysis tasks, effectively transferring potential information. Extensive experiments demonstrate the superiority of our approaches, with MCTI outperforming state-of-the-art frameworks on three public benchmarks. \href{https://github.com/jsh0792/MCTI}{https://github.com/jsh0792/MCTI}.
H2ASeg: Hierarchical Adaptive Interaction and Weighting Network for Tumor Segmentation in PET/CT Images
Mar 28, 2024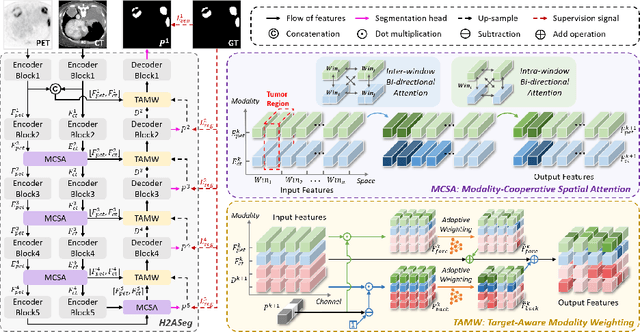
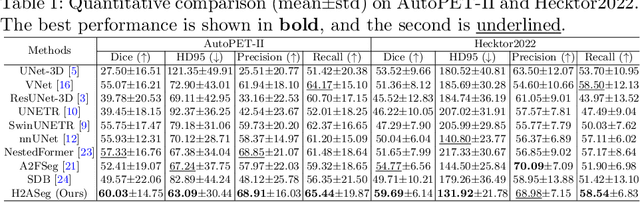
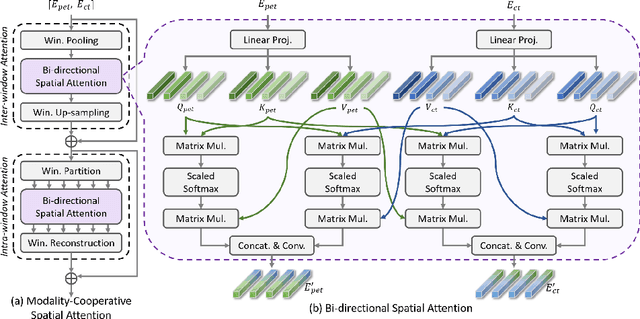
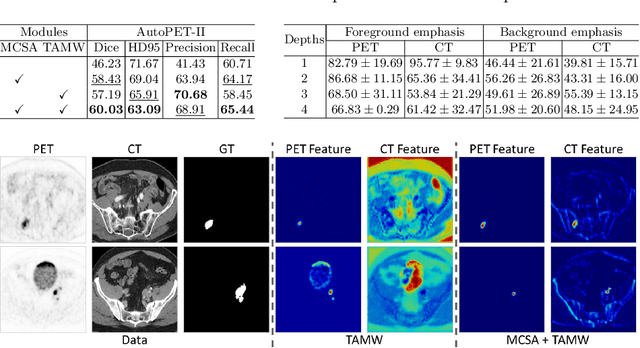
Abstract:Positron emission tomography (PET) combined with computed tomography (CT) imaging is routinely used in cancer diagnosis and prognosis by providing complementary information. Automatically segmenting tumors in PET/CT images can significantly improve examination efficiency. Traditional multi-modal segmentation solutions mainly rely on concatenation operations for modality fusion, which fail to effectively model the non-linear dependencies between PET and CT modalities. Recent studies have investigated various approaches to optimize the fusion of modality-specific features for enhancing joint representations. However, modality-specific encoders used in these methods operate independently, inadequately leveraging the synergistic relationships inherent in PET and CT modalities, for example, the complementarity between semantics and structure. To address these issues, we propose a Hierarchical Adaptive Interaction and Weighting Network termed H2ASeg to explore the intrinsic cross-modal correlations and transfer potential complementary information. Specifically, we design a Modality-Cooperative Spatial Attention (MCSA) module that performs intra- and inter-modal interactions globally and locally. Additionally, a Target-Aware Modality Weighting (TAMW) module is developed to highlight tumor-related features within multi-modal features, thereby refining tumor segmentation. By embedding these modules across different layers, H2ASeg can hierarchically model cross-modal correlations, enabling a nuanced understanding of both semantic and structural tumor features. Extensive experiments demonstrate the superiority of H2ASeg, outperforming state-of-the-art methods on AutoPet-II and Hecktor2022 benchmarks. The code is released at https://github.com/JinPLu/H2ASeg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge