Zhengyu Gan
Multimodal Cross-Task Interaction for Survival Analysis in Whole Slide Pathological Images
Jun 25, 2024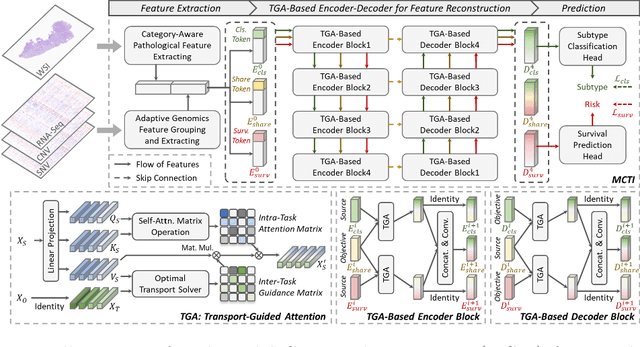
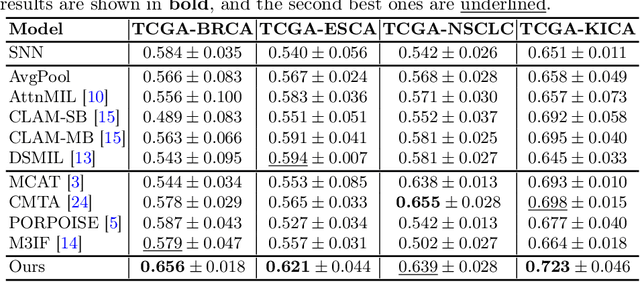
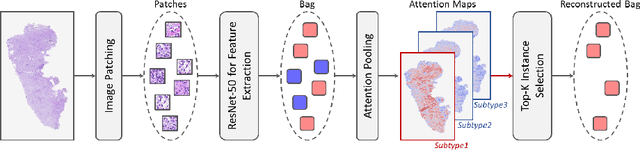

Abstract:Survival prediction, utilizing pathological images and genomic profiles, is increasingly important in cancer analysis and prognosis. Despite significant progress, precise survival analysis still faces two main challenges: (1) The massive pixels contained in whole slide images (WSIs) complicate the process of pathological images, making it difficult to generate an effective representation of the tumor microenvironment (TME). (2) Existing multimodal methods often rely on alignment strategies to integrate complementary information, which may lead to information loss due to the inherent heterogeneity between pathology and genes. In this paper, we propose a Multimodal Cross-Task Interaction (MCTI) framework to explore the intrinsic correlations between subtype classification and survival analysis tasks. Specifically, to capture TME-related features in WSIs, we leverage the subtype classification task to mine tumor regions. Simultaneously, multi-head attention mechanisms are applied in genomic feature extraction, adaptively performing genes grouping to obtain task-related genomic embedding. With the joint representation of pathological images and genomic data, we further introduce a Transport-Guided Attention (TGA) module that uses optimal transport theory to model the correlation between subtype classification and survival analysis tasks, effectively transferring potential information. Extensive experiments demonstrate the superiority of our approaches, with MCTI outperforming state-of-the-art frameworks on three public benchmarks. \href{https://github.com/jsh0792/MCTI}{https://github.com/jsh0792/MCTI}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge