Yeh-Chi Lo
A Deep Regression Model for Seed Identification in Prostate Brachytherapy
Jun 24, 2019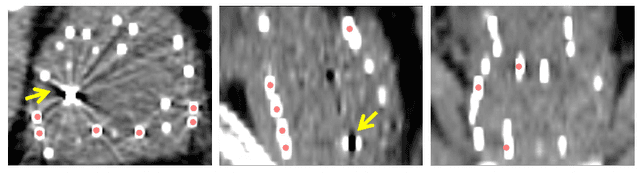
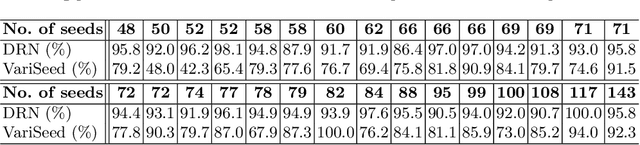
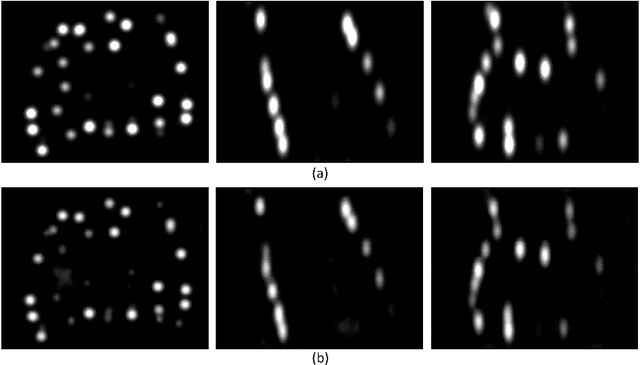
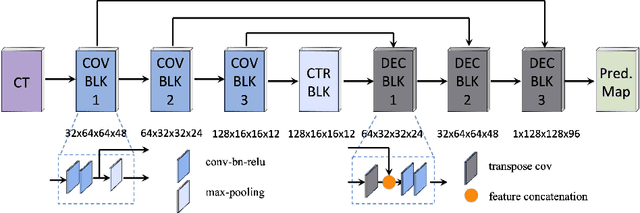
Abstract:Post-implant dosimetry (PID) is an essential step of prostate brachytherapy that utilizes CT to image the prostate and allow the location and dose distribution of the radioactive seeds to be directly related to the actual prostate. However, it it a very challenging task to identify these seeds in CT images due to the severe metal artifacts and high-overlapped appearance when multiple seeds clustered together. In this paper, we propose an automatic and efficient algorithm based on 3D deep fully convolutional network for identifying implanted seeds in CT images. Our method models the seed localization task as a supervised regression problem that projects the input CT image to a map where each element represents the probability that the corresponding input voxel belongs to a seed. This deep regression model significantly suppresses image artifacts and makes the post-processing much easier and more controllable. The proposed method is validated on a large clinical database with 7820 seeds in 100 patients, in which 5534 seeds from 70 patients were used for model training and validation. Our method correctly detected 2150 of 2286 (94.1%) seeds in the 30 testing patients, yielding 16% improvement as compared to a widely-used commercial seed finder software (VariSeed, Varian, Palo Alto, CA).
Improving Dermoscopic Image Segmentation with Enhanced Convolutional-Deconvolutional Networks
Sep 28, 2017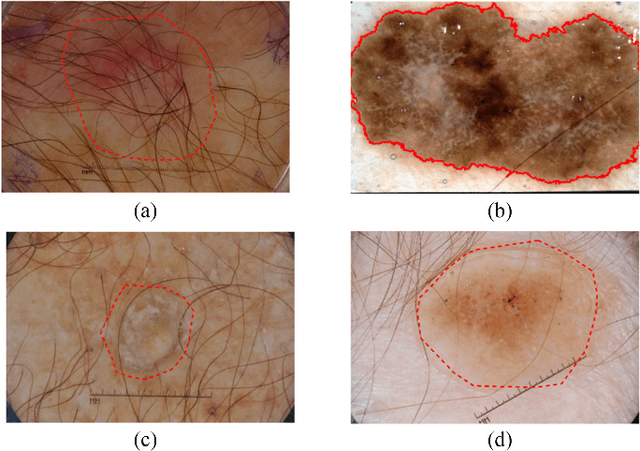
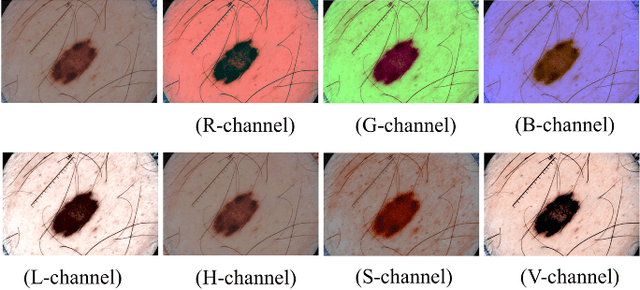
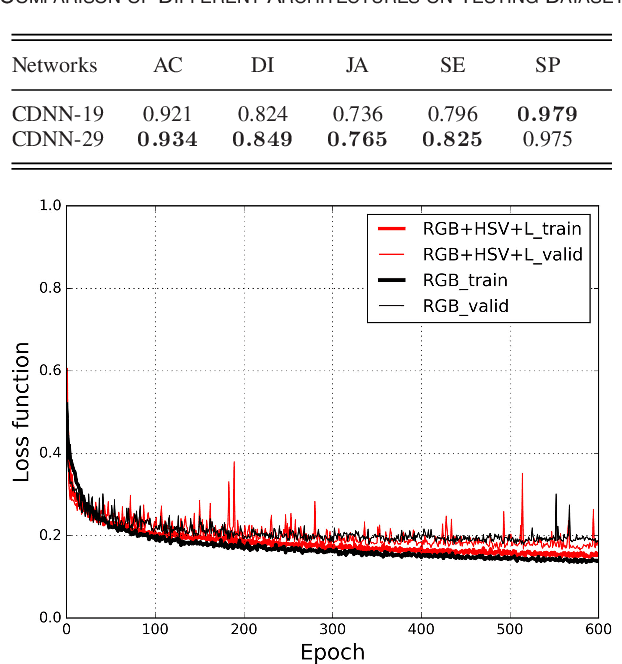
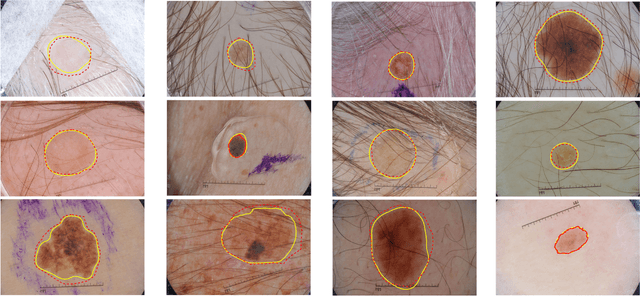
Abstract:Automatic skin lesion segmentation on dermoscopic images is an essential step in computer-aided diagnosis of melanoma. However, this task is challenging due to significant variations of lesion appearances across different patients. This challenge is further exacerbated when dealing with a large amount of image data. In this paper, we extended our previous work by developing a deeper network architecture with smaller kernels to enhance its discriminant capacity. In addition, we explicitly included color information from multiple color spaces to facilitate network training and thus to further improve the segmentation performance. We extensively evaluated our method on the ISBI 2017 skin lesion segmentation challenge. By training with the 2000 challenge training images, our method achieved an average Jaccard Index (JA) of 0.765 on the 600 challenge testing images, which ranked itself in the first place in the challenge
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge