Xiangzhe Kong
From Macro to Micro: Benchmarking Microscopic Spatial Intelligence on Molecules via Vision-Language Models
Dec 12, 2025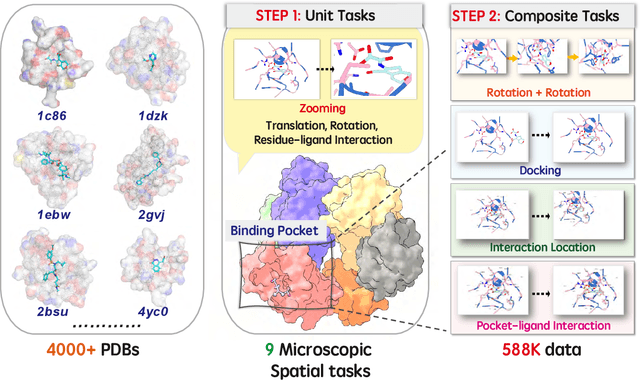
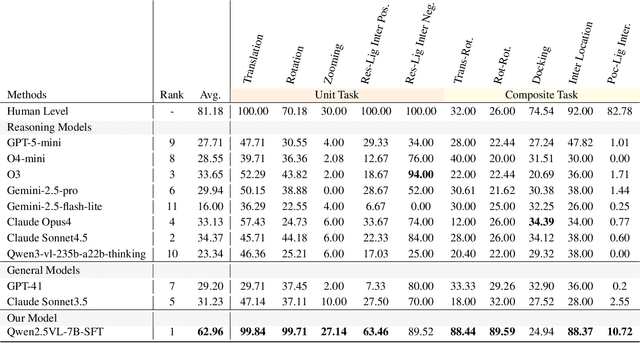
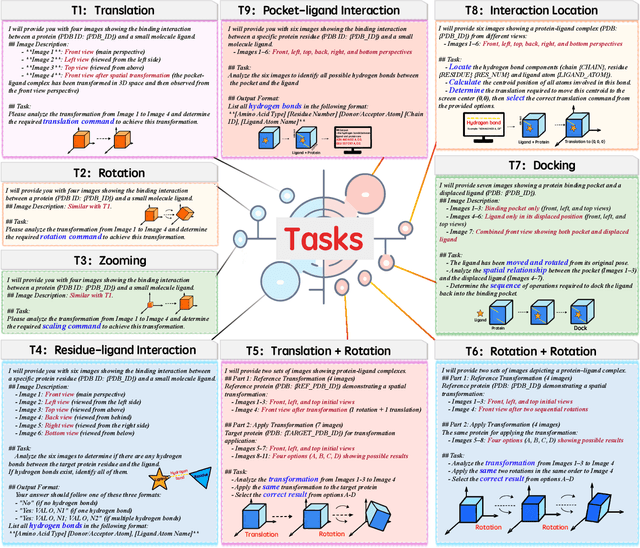
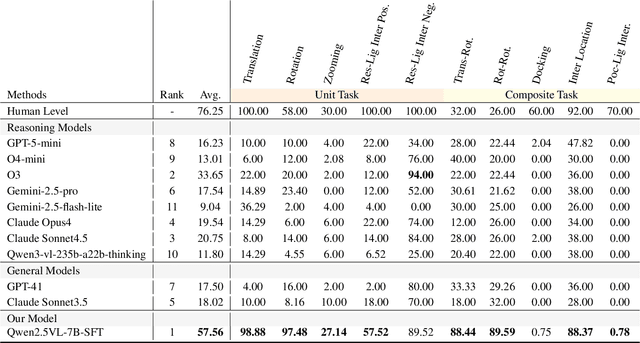
Abstract:This paper introduces the concept of Microscopic Spatial Intelligence (MiSI), the capability to perceive and reason about the spatial relationships of invisible microscopic entities, which is fundamental to scientific discovery. To assess the potential of Vision-Language Models (VLMs) in this domain, we propose a systematic benchmark framework MiSI-Bench. This framework features over 163,000 question-answer pairs and 587,000 images derived from approximately 4,000 molecular structures, covering nine complementary tasks that evaluate abilities ranging from elementary spatial transformations to complex relational identifications. Experimental results reveal that current state-of-the-art VLMs perform significantly below human level on this benchmark. However, a fine-tuned 7B model demonstrates substantial potential, even surpassing humans in spatial transformation tasks, while its poor performance in scientifically-grounded tasks like hydrogen bond recognition underscores the necessity of integrating explicit domain knowledge for progress toward scientific AGI. The datasets are available at https://huggingface.co/datasets/zongzhao/MiSI-bench.
Peptide2Mol: A Diffusion Model for Generating Small Molecules as Peptide Mimics for Targeted Protein Binding
Nov 07, 2025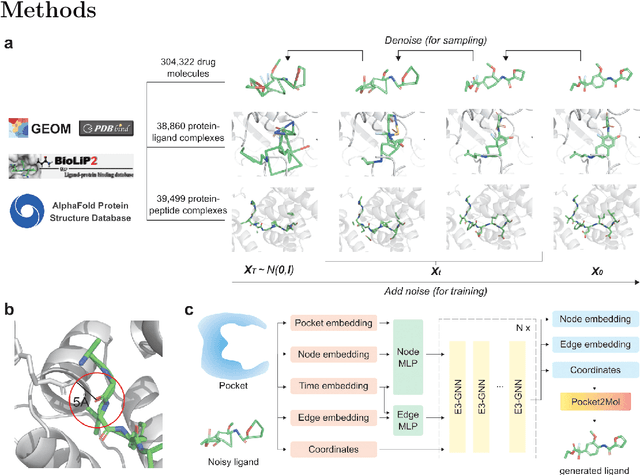
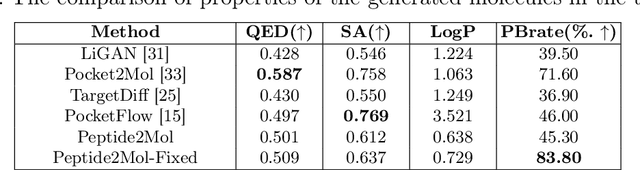
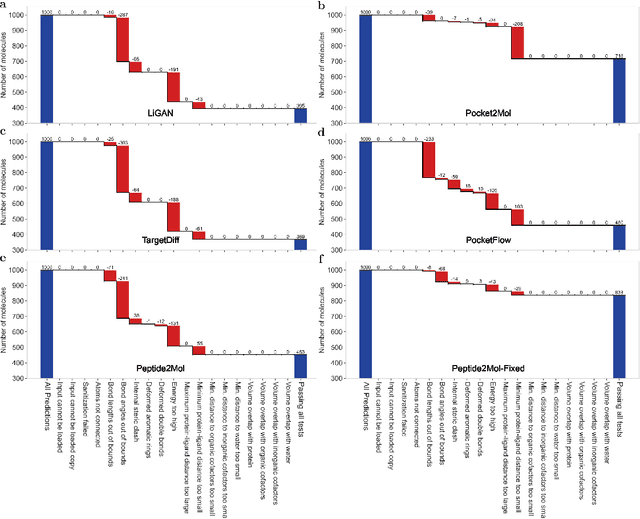
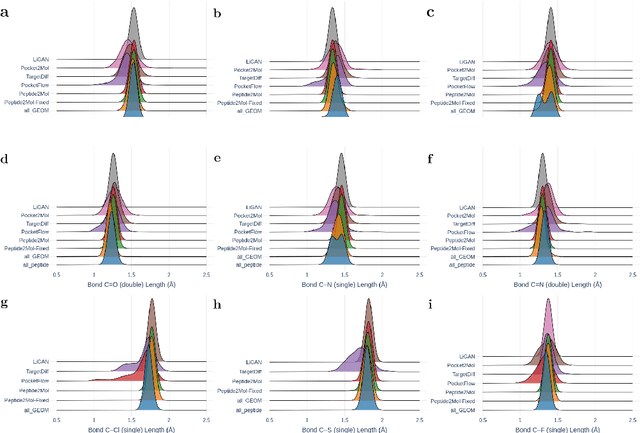
Abstract:Structure-based drug design has seen significant advancements with the integration of artificial intelligence (AI), particularly in the generation of hit and lead compounds. However, most AI-driven approaches neglect the importance of endogenous protein interactions with peptides, which may result in suboptimal molecule designs. In this work, we present Peptide2Mol, an E(3)-equivariant graph neural network diffusion model that generates small molecules by referencing both the original peptide binders and their surrounding protein pocket environments. Trained on large datasets and leveraging sophisticated modeling techniques, Peptide2Mol not only achieves state-of-the-art performance in non-autoregressive generative tasks, but also produces molecules with similarity to the original peptide binder. Additionally, the model allows for molecule optimization and peptidomimetic design through a partial diffusion process. Our results highlight Peptide2Mol as an effective deep generative model for generating and optimizing bioactive small molecules from protein binding pockets.
UniMoMo: Unified Generative Modeling of 3D Molecules for De Novo Binder Design
Mar 25, 2025Abstract:The design of target-specific molecules such as small molecules, peptides, and antibodies is vital for biological research and drug discovery. Existing generative methods are restricted to single-domain molecules, failing to address versatile therapeutic needs or utilize cross-domain transferability to enhance model performance. In this paper, we introduce Unified generative Modeling of 3D Molecules (UniMoMo), the first framework capable of designing binders of multiple molecular domains using a single model. In particular, UniMoMo unifies the representations of different molecules as graphs of blocks, where each block corresponds to either a standard amino acid or a molecular fragment. Based on these unified representations, UniMoMo utilizes a geometric latent diffusion model for 3D molecular generation, featuring an iterative full-atom autoencoder to compress blocks into latent space points, followed by an E(3)-equivariant diffusion process. Extensive benchmarks across peptides, antibodies, and small molecules demonstrate the superiority of our unified framework over existing domain-specific models, highlighting the benefits of multi-domain training.
A Survey of Geometric Graph Neural Networks: Data Structures, Models and Applications
Mar 01, 2024Abstract:Geometric graph is a special kind of graph with geometric features, which is vital to model many scientific problems. Unlike generic graphs, geometric graphs often exhibit physical symmetries of translations, rotations, and reflections, making them ineffectively processed by current Graph Neural Networks (GNNs). To tackle this issue, researchers proposed a variety of Geometric Graph Neural Networks equipped with invariant/equivariant properties to better characterize the geometry and topology of geometric graphs. Given the current progress in this field, it is imperative to conduct a comprehensive survey of data structures, models, and applications related to geometric GNNs. In this paper, based on the necessary but concise mathematical preliminaries, we provide a unified view of existing models from the geometric message passing perspective. Additionally, we summarize the applications as well as the related datasets to facilitate later research for methodology development and experimental evaluation. We also discuss the challenges and future potential directions of Geometric GNNs at the end of this survey.
Equivariant Pretrained Transformer for Unified Geometric Learning on Multi-Domain 3D Molecules
Feb 20, 2024Abstract:Pretraining on a large number of unlabeled 3D molecules has showcased superiority in various scientific applications. However, prior efforts typically focus on pretraining models on a specific domain, either proteins or small molecules, missing the opportunity to leverage the cross-domain knowledge. To mitigate this gap, we introduce Equivariant Pretrained Transformer (EPT), a novel pretraining framework designed to harmonize the geometric learning of small molecules and proteins. To be specific, EPT unifies the geometric modeling of multi-domain molecules via the block-enhanced representation that can attend a broader context of each atom. Upon transformer framework, EPT is further enhanced with E(3) equivariance to facilitate the accurate representation of 3D structures. Another key innovation of EPT is its block-level pretraining task, which allows for joint pretraining on datasets comprising both small molecules and proteins. Experimental evaluations on a diverse group of benchmarks, including ligand binding affinity prediction, molecular property prediction, and protein property prediction, show that EPT significantly outperforms previous SOTA methods for affinity prediction, and achieves the best or comparable performance with existing domain-specific pretraining models for other tasks.
Generalist Equivariant Transformer Towards 3D Molecular Interaction Learning
Jun 05, 2023Abstract:Many processes in biology and drug discovery involve various 3D interactions between different molecules, such as protein and protein, protein and small molecule, etc. Designing a generalist model to learn universal molecular interactions is valuable yet challenging, given that different molecules are usually represented in different granularity. In this paper, we first propose to universally represent a 3D molecule as a geometric graph of sets, in contrast to conventional single-level representations. Upon the proposed unified representation, we then propose a Generalist Equivariant Transformer (GET) to effectively capture both sparse block-level and dense atom-level interactions. To be specific, GET consists of a bilevel attention module, a feed-forward module and a layer normalization module, where, notably, each module is E(3) equivariant to meet the symmetry of 3D world. Extensive experiments on the prediction of protein-protein affinity, ligand binding affinity, and ligand efficacy prediction verify the effectiveness of our proposed method against existing methods, and reveal its potential to learn transferable knowledge across different domains and different tasks.
End-to-End Full-Atom Antibody Design
Feb 15, 2023



Abstract:Antibody design is an essential yet challenging task in various domains like therapeutics and biology. There are two major defects in current learning-based methods: 1) tackling only a certain subtask of the whole antibody design pipeline, making them suboptimal or resource-intensive. 2) omitting either the framework regions or side chains, thus incapable of capturing the full-atom geometry. To address these pitfalls, we propose dynamic Multi-channel Equivariant grAph Network (dyMEAN), an end-to-end full-atom model for E(3)-equivariant antibody design given the epitope and the incomplete sequence of the antibody. Specifically, we first explore structural initialization as a knowledgeable guess of the antibody structure and then propose shadow paratope to bridge the epitope-antibody connections. Both 1D sequences and 3D structures are updated via an adaptive multi-channel equivariant encoder that is able to process protein residues of variable sizes when considering full atoms. Finally, the updated antibody is docked to the epitope via the alignment of the shadow paratope. Experiments on epitope-binding CDR-H3 design, complex structure prediction, and affinity optimization demonstrate the superiority of our end-to-end framework and full-atom modeling.
Conditional Antibody Design as 3D Equivariant Graph Translation
Aug 12, 2022



Abstract:Antibody design is valuable for therapeutic usage and biological research. Existing deep-learning-based methods encounter several key issues: 1) incomplete context for Complementarity-Determining Regions (CDRs) generation; 2) incapable of capturing the entire 3D geometry of the input structure; 3) inefficient prediction of the CDR sequences in an autoregressive manner. In this paper, we propose Multi-channel Equivariant Attention Network (MEAN), an end-to-end model that is able to co-design 1D sequences and 3D structures of CDRs. To be specific, MEAN formulates antibody design as a conditional graph translation problem by importing extra components including the target antigen and the light chain of the antibody. Then, MEAN resorts to E(3)-equivariant message passing along with a proposed attention mechanism to better capture the geometrical correlation between different components. Finally, it outputs both the 1D sequences and 3D structure via a multi-round progressive full-shot scheme, which enjoys more efficiency against previous autoregressive approaches. Our method significantly surpasses state-of-the-art models in sequence and structure modeling, antigen-binding antibody design, and binding affinity optimization. Specifically, the relative improvement to baselines is about 22% in antigen-binding CDR design and 34% for affinity optimization.
GraphPiece: Efficiently Generating High-Quality Molecular Graph with Substructures
Jun 29, 2021



Abstract:Molecular graph generation is a fundamental but challenging task in various applications such as drug discovery and material science, which requires generating valid molecules with desired properties. Auto-regressive models, which usually construct graphs following sequential actions of adding nodes and edges at the atom-level, have made rapid progress in recent years. However, these atom-level models ignore high-frequency subgraphs that not only capture the regularities of atomic combination in molecules but also are often related to desired chemical properties. In this paper, we propose a method to automatically discover such common substructures, which we call {\em graph pieces}, from given molecular graphs. Based on graph pieces, we leverage a variational autoencoder to generate molecules in two phases: piece-level graph generation followed by bond completion. Experiments show that our graph piece variational autoencoder achieves better performance over state-of-the-art baselines on property optimization and constrained property optimization tasks with higher computational efficiency.
Stylized Story Generation with Style-Guided Planning
May 19, 2021



Abstract:Current storytelling systems focus more ongenerating stories with coherent plots regard-less of the narration style, which is impor-tant for controllable text generation. There-fore, we propose a new task, stylized story gen-eration, namely generating stories with speci-fied style given a leading context. To tacklethe problem, we propose a novel generationmodel that first plans the stylized keywordsand then generates the whole story with theguidance of the keywords. Besides, we pro-pose two automatic metrics to evaluate theconsistency between the generated story andthe specified style. Experiments demonstratesthat our model can controllably generateemo-tion-driven orevent-driven stories based onthe ROCStories dataset (Mostafazadeh et al.,2016). Our study presents insights for stylizedstory generation in further research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge