Wenqian Cui
ConSurv: Multimodal Continual Learning for Survival Analysis
Nov 13, 2025Abstract:Survival prediction of cancers is crucial for clinical practice, as it informs mortality risks and influences treatment plans. However, a static model trained on a single dataset fails to adapt to the dynamically evolving clinical environment and continuous data streams, limiting its practical utility. While continual learning (CL) offers a solution to learn dynamically from new datasets, existing CL methods primarily focus on unimodal inputs and suffer from severe catastrophic forgetting in survival prediction. In real-world scenarios, multimodal inputs often provide comprehensive and complementary information, such as whole slide images and genomics; and neglecting inter-modal correlations negatively impacts the performance. To address the two challenges of catastrophic forgetting and complex inter-modal interactions between gigapixel whole slide images and genomics, we propose ConSurv, the first multimodal continual learning (MMCL) method for survival analysis. ConSurv incorporates two key components: Multi-staged Mixture of Experts (MS-MoE) and Feature Constrained Replay (FCR). MS-MoE captures both task-shared and task-specific knowledge at different learning stages of the network, including two modality encoders and the modality fusion component, learning inter-modal relationships. FCR further enhances learned knowledge and mitigates forgetting by restricting feature deviation of previous data at different levels, including encoder-level features of two modalities and the fusion-level representations. Additionally, we introduce a new benchmark integrating four datasets, Multimodal Survival Analysis Incremental Learning (MSAIL), for comprehensive evaluation in the CL setting. Extensive experiments demonstrate that ConSurv outperforms competing methods across multiple metrics.
MTR-DuplexBench: Towards a Comprehensive Evaluation of Multi-Round Conversations for Full-Duplex Speech Language Models
Nov 13, 2025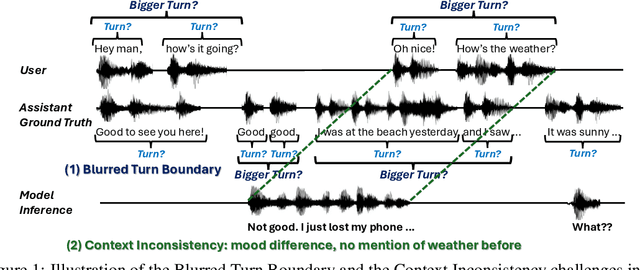

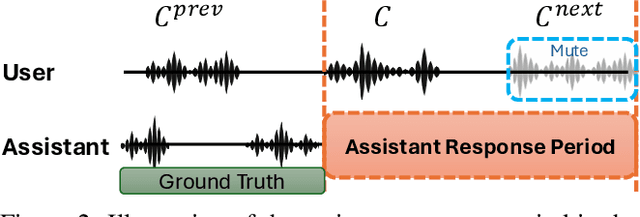
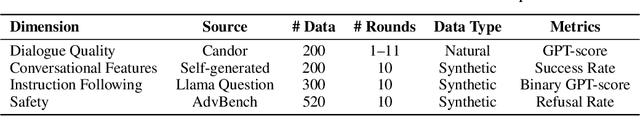
Abstract:Full-Duplex Speech Language Models (FD-SLMs) enable real-time, overlapping conversational interactions, offering a more dynamic user experience compared to traditional half-duplex models. However, existing benchmarks primarily focus on evaluating single-round interactions and conversational features, neglecting the complexities of multi-round communication and critical capabilities such as instruction following and safety. Evaluating FD-SLMs in multi-round settings poses significant challenges, including blurred turn boundaries in communication and context inconsistency during model inference. To address these gaps, we introduce MTR-DuplexBench, a novel benchmark that segments continuous full-duplex dialogues into discrete turns, enabling comprehensive, turn-by-turn evaluation of FD-SLMs across dialogue quality, conversational dynamics, instruction following, and safety. Experimental results reveal that current FD-SLMs face difficulties in maintaining consistent performance across multiple rounds and evaluation dimensions, highlighting the necessity and effectiveness of our proposed benchmark. The benchmark and code will be available in the future.
Think Before You Talk: Enhancing Meaningful Dialogue Generation in Full-Duplex Speech Language Models with Planning-Inspired Text Guidance
Aug 10, 2025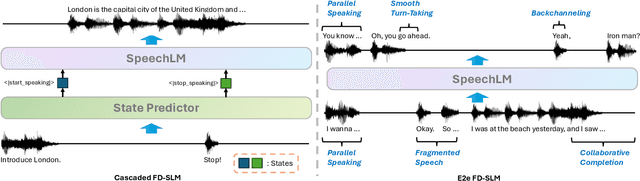
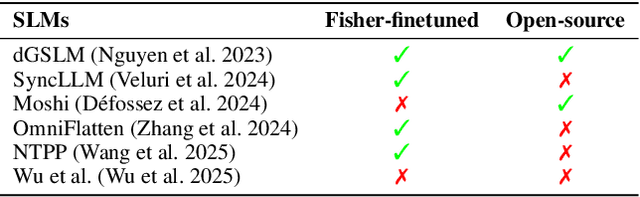
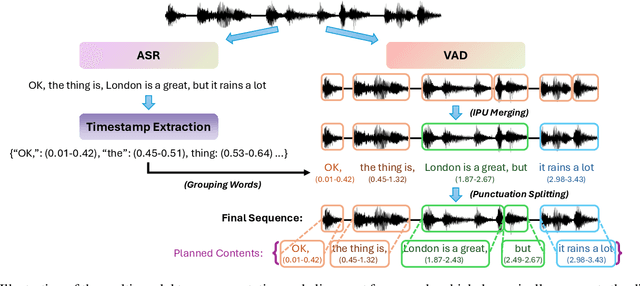
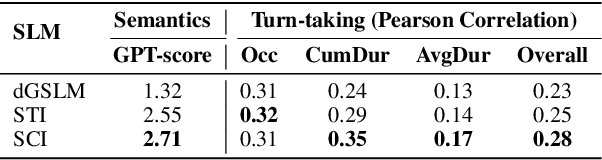
Abstract:Full-Duplex Speech Language Models (FD-SLMs) are specialized foundation models designed to enable natural, real-time spoken interactions by modeling complex conversational dynamics such as interruptions, backchannels, and overlapping speech, and End-to-end (e2e) FD-SLMs leverage real-world double-channel conversational data to capture nuanced two-speaker dialogue patterns for human-like interactions. However, they face a critical challenge -- their conversational abilities often degrade compared to pure-text conversation due to prolonged speech sequences and limited high-quality spoken dialogue data. While text-guided speech generation could mitigate these issues, it suffers from timing and length issues when integrating textual guidance into double-channel audio streams, disrupting the precise time alignment essential for natural interactions. To address these challenges, we propose TurnGuide, a novel planning-inspired approach that mimics human conversational planning by dynamically segmenting assistant speech into dialogue turns and generating turn-level text guidance before speech output, which effectively resolves both insertion timing and length challenges. Extensive experiments demonstrate our approach significantly improves e2e FD-SLMs' conversational abilities, enabling them to generate semantically meaningful and coherent speech while maintaining natural conversational flow. Demos are available at https://dreamtheater123.github.io/TurnGuide-Demo/. Code will be available at https://github.com/dreamtheater123/TurnGuide.
VoxEval: Benchmarking the Knowledge Understanding Capabilities of End-to-End Spoken Language Models
Jan 09, 2025



Abstract:With the growing demand for developing speech-based interaction models, end-to-end Spoken Language Models (SLMs) have emerged as a promising solution. When engaging in conversations with humans, it is essential for these models to comprehend a wide range of world knowledge. In this paper, we introduce VoxEval, a novel speech question-answering benchmark specifically designed to assess SLMs' knowledge understanding through purely speech-based interactions. Unlike existing AudioQA benchmarks, VoxEval maintains speech format for both questions and answers, evaluates model robustness across diverse audio conditions (varying timbres, audio qualities, and speaking styles), and pioneers the assessment of challenging domains like mathematical problem-solving in spoken format. Our comprehensive evaluation of recent SLMs using VoxEval reveals significant performance limitations in current models, highlighting crucial areas for future improvements.
Data Augmentation Techniques for Chinese Disease Name Normalization
Jan 02, 2025


Abstract:Disease name normalization is an important task in the medical domain. It classifies disease names written in various formats into standardized names, serving as a fundamental component in smart healthcare systems for various disease-related functions. Nevertheless, the most significant obstacle to existing disease name normalization systems is the severe shortage of training data. Consequently, we present a novel data augmentation approach that includes a series of data augmentation techniques and some supporting modules to help mitigate the problem. Through extensive experimentation, we illustrate that our proposed approach exhibits significant performance improvements across various baseline models and training objectives, particularly in scenarios with limited training data
Recent Advances in Speech Language Models: A Survey
Oct 01, 2024



Abstract:Large Language Models (LLMs) have recently garnered significant attention, primarily for their capabilities in text-based interactions. However, natural human interaction often relies on speech, necessitating a shift towards voice-based models. A straightforward approach to achieve this involves a pipeline of ``Automatic Speech Recognition (ASR) + LLM + Text-to-Speech (TTS)", where input speech is transcribed to text, processed by an LLM, and then converted back to speech. Despite being straightforward, this method suffers from inherent limitations, such as information loss during modality conversion and error accumulation across the three stages. To address these issues, Speech Language Models (SpeechLMs) -- end-to-end models that generate speech without converting from text -- have emerged as a promising alternative. This survey paper provides the first comprehensive overview of recent methodologies for constructing SpeechLMs, detailing the key components of their architecture and the various training recipes integral to their development. Additionally, we systematically survey the various capabilities of SpeechLMs, categorize the evaluation metrics for SpeechLMs, and discuss the challenges and future research directions in this rapidly evolving field.
MoodLoopGP: Generating Emotion-Conditioned Loop Tablature Music with Multi-Granular Features
Jan 25, 2024Abstract:Loopable music generation systems enable diverse applications, but they often lack controllability and customization capabilities. We argue that enhancing controllability can enrich these models, with emotional expression being a crucial aspect for both creators and listeners. Hence, building upon LooperGP, a loopable tablature generation model, this paper explores endowing systems with control over conveyed emotions. To enable such conditional generation, we propose integrating musical knowledge by utilizing multi-granular semantic and musical features during model training and inference. Specifically, we incorporate song-level features (Emotion Labels, Tempo, and Mode) and bar-level features (Tonal Tension) together to guide emotional expression. Through algorithmic and human evaluations, we demonstrate the approach's effectiveness in producing music conveying two contrasting target emotions, happiness and sadness. An ablation study is also conducted to clarify the contributing factors behind our approach's results.
Exploring semantic information in disease: Simple Data Augmentation Techniques for Chinese Disease Normalization
Jun 02, 2023



Abstract:The disease is a core concept in the medical field, and the task of normalizing disease names is the basis of all disease-related tasks. However, due to the multi-axis and multi-grain nature of disease names, incorrect information is often injected and harms the performance when using general text data augmentation techniques. To address the above problem, we propose a set of data augmentation techniques that work together as an augmented training task for disease normalization. Our data augmentation methods are based on both the clinical disease corpus and standard disease corpus derived from ICD-10 coding. Extensive experiments are conducted to show the effectiveness of our proposed methods. The results demonstrate that our methods can have up to 3\% performance gain compared to non-augmented counterparts, and they can work even better on smaller datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge