Wanru Zhuang
Towards Cross-Modal Text-Molecule Retrieval with Better Modality Alignment
Oct 31, 2024Abstract:Cross-modal text-molecule retrieval model aims to learn a shared feature space of the text and molecule modalities for accurate similarity calculation, which facilitates the rapid screening of molecules with specific properties and activities in drug design. However, previous works have two main defects. First, they are inadequate in capturing modality-shared features considering the significant gap between text sequences and molecule graphs. Second, they mainly rely on contrastive learning and adversarial training for cross-modality alignment, both of which mainly focus on the first-order similarity, ignoring the second-order similarity that can capture more structural information in the embedding space. To address these issues, we propose a novel cross-modal text-molecule retrieval model with two-fold improvements. Specifically, on the top of two modality-specific encoders, we stack a memory bank based feature projector that contain learnable memory vectors to extract modality-shared features better. More importantly, during the model training, we calculate four kinds of similarity distributions (text-to-text, text-to-molecule, molecule-to-molecule, and molecule-to-text similarity distributions) for each instance, and then minimize the distance between these similarity distributions (namely second-order similarity losses) to enhance cross-modal alignment. Experimental results and analysis strongly demonstrate the effectiveness of our model. Particularly, our model achieves SOTA performance, outperforming the previously-reported best result by 6.4%.
Empowering Backbone Models for Visual Text Generation with Input Granularity Control and Glyph-Aware Training
Oct 06, 2024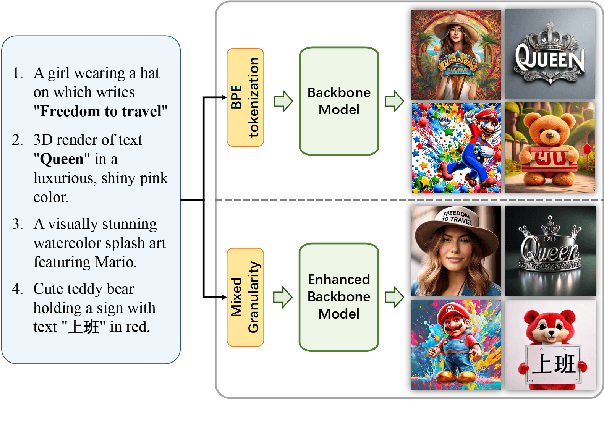
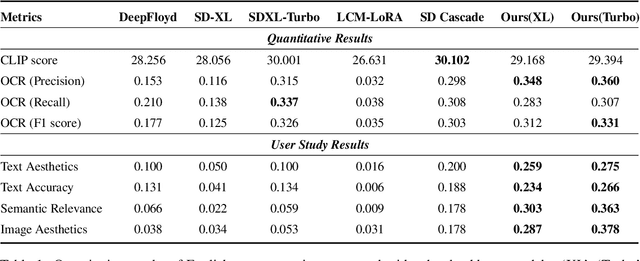
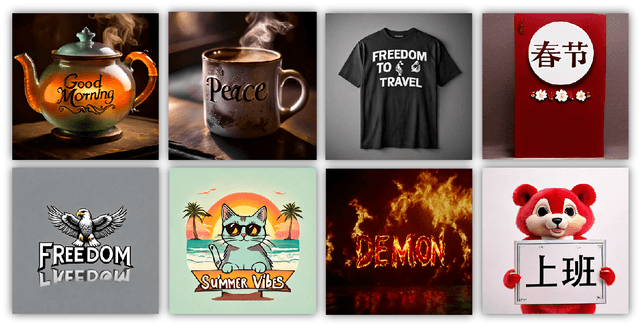

Abstract:Diffusion-based text-to-image models have demonstrated impressive achievements in diversity and aesthetics but struggle to generate images with legible visual texts. Existing backbone models have limitations such as misspelling, failing to generate texts, and lack of support for Chinese text, but their development shows promising potential. In this paper, we propose a series of methods, aiming to empower backbone models to generate visual texts in English and Chinese. We first conduct a preliminary study revealing that Byte Pair Encoding (BPE) tokenization and the insufficient learning of cross-attention modules restrict the performance of the backbone models. Based on these observations, we make the following improvements: (1) We design a mixed granularity input strategy to provide more suitable text representations; (2) We propose to augment the conventional training objective with three glyph-aware training losses, which enhance the learning of cross-attention modules and encourage the model to focus on visual texts. Through experiments, we demonstrate that our methods can effectively empower backbone models to generate semantic relevant, aesthetically appealing, and accurate visual text images, while maintaining their fundamental image generation quality.
Multi-level protein pre-training with Vabs-Net
Feb 05, 2024Abstract:In recent years, there has been a surge in the development of 3D structure-based pre-trained protein models, representing a significant advancement over pre-trained protein language models in various downstream tasks. However, most existing structure-based pre-trained models primarily focus on the residue level, i.e., alpha carbon atoms, while ignoring other atoms like side chain atoms. We argue that modeling proteins at both residue and atom levels is important since the side chain atoms can also be crucial for numerous downstream tasks, for example, molecular docking. Nevertheless, we find that naively combining residue and atom information during pre-training typically fails. We identify a key reason is the information leakage caused by the inclusion of atom structure in the input, which renders residue-level pre-training tasks trivial and results in insufficiently expressive residue representations. To address this issue, we introduce a span mask pre-training strategy on 3D protein chains to learn meaningful representations of both residues and atoms. This leads to a simple yet effective approach to learning protein representation suitable for diverse downstream tasks. Extensive experimental results on binding site prediction and function prediction tasks demonstrate our proposed pre-training approach significantly outperforms other methods. Our code will be made public.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge