Tuan Le
Byzantine Resilient Federated Multi-Task Representation Learning
Mar 24, 2025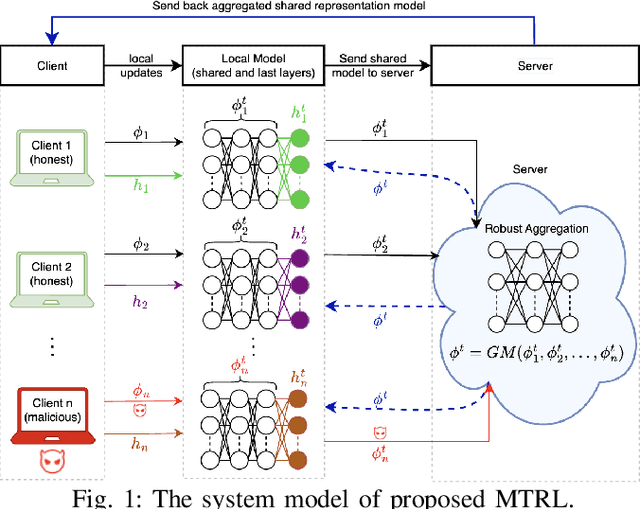
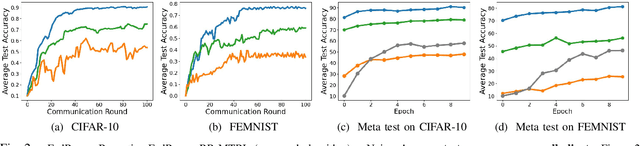
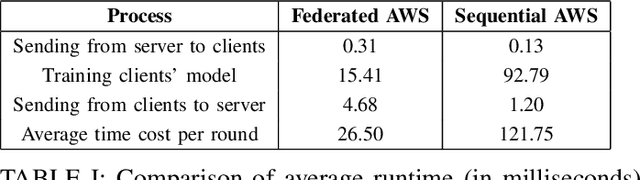
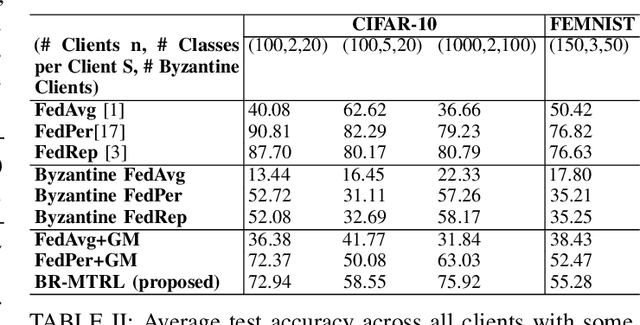
Abstract:In this paper, we propose BR-MTRL, a Byzantine-resilient multi-task representation learning framework that handles faulty or malicious agents. Our approach leverages representation learning through a shared neural network model, where all clients share fixed layers, except for a client-specific final layer. This structure captures shared features among clients while enabling individual adaptation, making it a promising approach for leveraging client data and computational power in heterogeneous federated settings to learn personalized models. To learn the model, we employ an alternating gradient descent strategy: each client optimizes its local model, updates its final layer, and sends estimates of the shared representation to a central server for aggregation. To defend against Byzantine agents, we employ geometric median aggregation for robust client-server communication. Our method enables personalized learning while maintaining resilience in distributed settings. We implemented the proposed alternating gradient descent algorithm in a federated testbed built using Amazon Web Services (AWS) platform and compared its performance with various benchmark algorithms and their variations. Through extensive experiments using real-world datasets, including CIFAR-10 and FEMINIST, we demonstrated the effectiveness and robustness of our approach and its transferability to new unseen clients with limited data, even in the presence of Byzantine adversaries.
Generative Modeling on Lie Groups via Euclidean Generalized Score Matching
Feb 04, 2025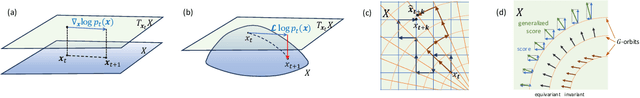
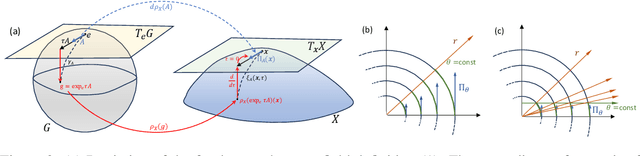
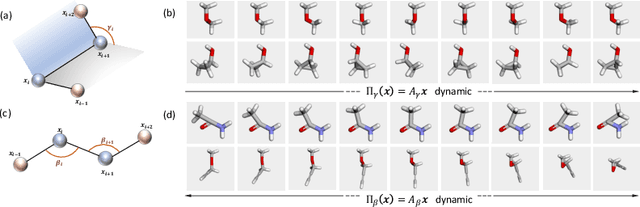
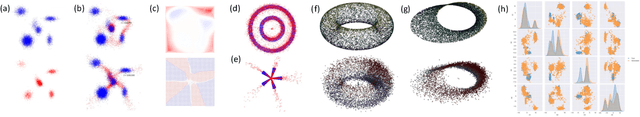
Abstract:We extend Euclidean score-based diffusion processes to generative modeling on Lie groups. Through the formalism of Generalized Score Matching, our approach yields a Langevin dynamics which decomposes as a direct sum of Lie algebra representations, enabling generative processes on Lie groups while operating in Euclidean space. Unlike equivariant models, which restrict the space of learnable functions by quotienting out group orbits, our method can model any target distribution on any (non-Abelian) Lie group. Standard score matching emerges as a special case of our framework when the Lie group is the translation group. We prove that our generalized generative processes arise as solutions to a new class of paired stochastic differential equations (SDEs), introduced here for the first time. We validate our approach through experiments on diverse data types, demonstrating its effectiveness in real-world applications such as SO(3)-guided molecular conformer generation and modeling ligand-specific global SE(3) transformations for molecular docking, showing improvement in comparison to Riemannian diffusion on the group itself. We show that an appropriate choice of Lie group enhances learning efficiency by reducing the effective dimensionality of the trajectory space and enables the modeling of transitions between complex data distributions. Additionally, we demonstrate the universality of our approach by deriving how it extends to flow matching.
PILOT: Equivariant diffusion for pocket conditioned de novo ligand generation with multi-objective guidance via importance sampling
May 23, 2024



Abstract:The generation of ligands that both are tailored to a given protein pocket and exhibit a range of desired chemical properties is a major challenge in structure-based drug design. Here, we propose an in-silico approach for the $\textit{de novo}$ generation of 3D ligand structures using the equivariant diffusion model PILOT, combining pocket conditioning with a large-scale pre-training and property guidance. Its multi-objective trajectory-based importance sampling strategy is designed to direct the model towards molecules that not only exhibit desired characteristics such as increased binding affinity for a given protein pocket but also maintains high synthetic accessibility. This ensures the practicality of sampled molecules, thus maximizing their potential for the drug discovery pipeline. PILOT significantly outperforms existing methods across various metrics on the common benchmark dataset CrossDocked2020. Moreover, we employ PILOT to generate novel ligands for unseen protein pockets from the Kinodata-3D dataset, which encompasses a substantial portion of the human kinome. The generated structures exhibit predicted $IC_{50}$ values indicative of potent biological activity, which highlights the potential of PILOT as a powerful tool for structure-based drug design.
A Chain-of-Thought Prompting Approach with LLMs for Evaluating Students' Formative Assessment Responses in Science
Mar 21, 2024



Abstract:This paper explores the use of large language models (LLMs) to score and explain short-answer assessments in K-12 science. While existing methods can score more structured math and computer science assessments, they often do not provide explanations for the scores. Our study focuses on employing GPT-4 for automated assessment in middle school Earth Science, combining few-shot and active learning with chain-of-thought reasoning. Using a human-in-the-loop approach, we successfully score and provide meaningful explanations for formative assessment responses. A systematic analysis of our method's pros and cons sheds light on the potential for human-in-the-loop techniques to enhance automated grading for open-ended science assessments.
Navigating the Design Space of Equivariant Diffusion-Based Generative Models for De Novo 3D Molecule Generation
Sep 29, 2023Abstract:Deep generative diffusion models are a promising avenue for de novo 3D molecular design in material science and drug discovery. However, their utility is still constrained by suboptimal performance with large molecular structures and limited training data. Addressing this gap, we explore the design space of E(3) equivariant diffusion models, focusing on previously blank spots. Our extensive comparative analysis evaluates the interplay between continuous and discrete state spaces. Out of this investigation, we introduce the EQGAT-diff model, which consistently surpasses the performance of established models on the QM9 and GEOM-Drugs datasets by a large margin. Distinctively, EQGAT-diff takes continuous atomic positions while chemical elements and bond types are categorical and employ a time-dependent loss weighting that significantly increases training convergence and the quality of generated samples. To further strengthen the applicability of diffusion models to limited training data, we examine the transferability of EQGAT-diff trained on the large PubChem3D dataset with implicit hydrogens to target distributions with explicit hydrogens. Fine-tuning EQGAT-diff for a couple of iterations further pushes state-of-the-art performance across datasets. We envision that our findings will find applications in structure-based drug design, where the accuracy of generative models for small datasets of complex molecules is critical.
Equivariant Graph Attention Networks for Molecular Property Prediction
Mar 02, 2022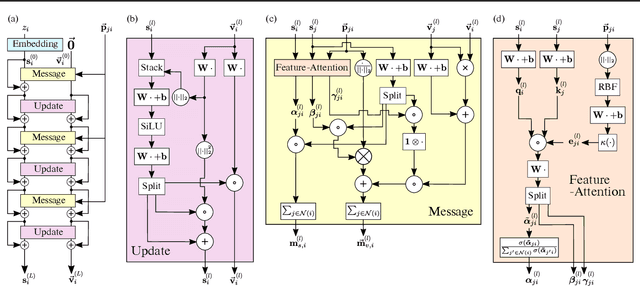
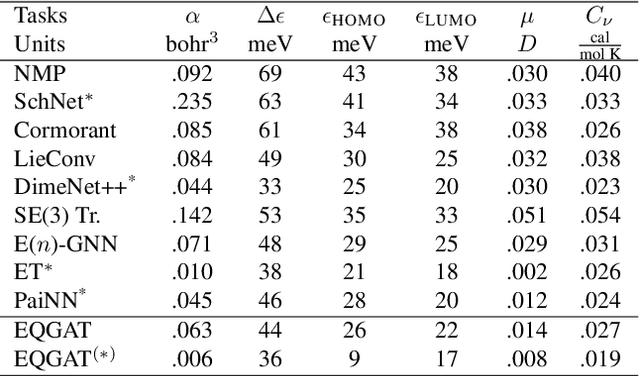
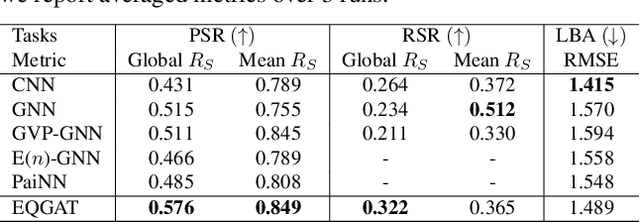
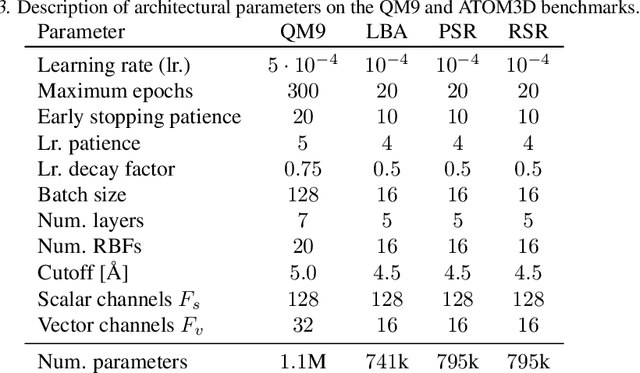
Abstract:Learning and reasoning about 3D molecular structures with varying size is an emerging and important challenge in machine learning and especially in drug discovery. Equivariant Graph Neural Networks (GNNs) can simultaneously leverage the geometric and relational detail of the problem domain and are known to learn expressive representations through the propagation of information between nodes leveraging higher-order representations to faithfully express the geometry of the data, such as directionality in their intermediate layers. In this work, we propose an equivariant GNN that operates with Cartesian coordinates to incorporate directionality and we implement a novel attention mechanism, acting as a content and spatial dependent filter when propagating information between nodes. We demonstrate the efficacy of our architecture on predicting quantum mechanical properties of small molecules and its benefit on problems that concern macromolecular structures such as protein complexes.
Unsupervised Learning of Group Invariant and Equivariant Representations
Feb 15, 2022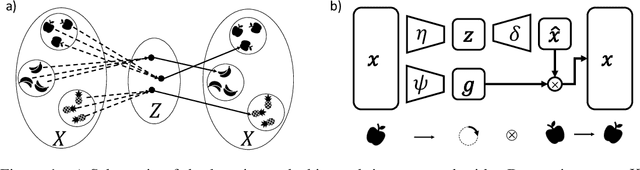
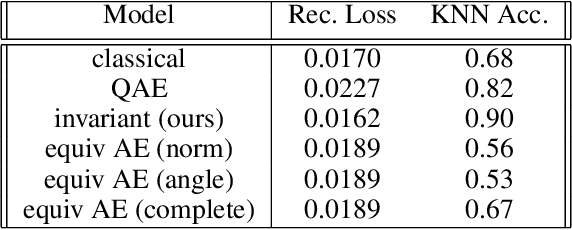

Abstract:Equivariant neural networks, whose hidden features transform according to representations of a group G acting on the data, exhibit training efficiency and an improved generalisation performance. In this work, we extend group invariant and equivariant representation learning to the field of unsupervised deep learning. We propose a general learning strategy based on an encoder-decoder framework in which the latent representation is disentangled in an invariant term and an equivariant group action component. The key idea is that the network learns the group action on the data space and thus is able to solve the reconstruction task from an invariant data representation, hence avoiding the necessity of ad-hoc group-specific implementations. We derive the necessary conditions on the equivariant encoder, and we present a construction valid for any G, both discrete and continuous. We describe explicitly our construction for rotations, translations and permutations. We test the validity and the robustness of our approach in a variety of experiments with diverse data types employing different network architectures.
Parameterized Hypercomplex Graph Neural Networks for Graph Classification
Mar 30, 2021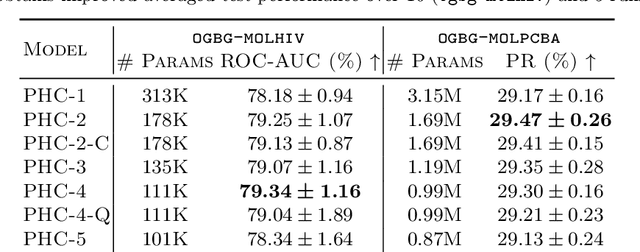
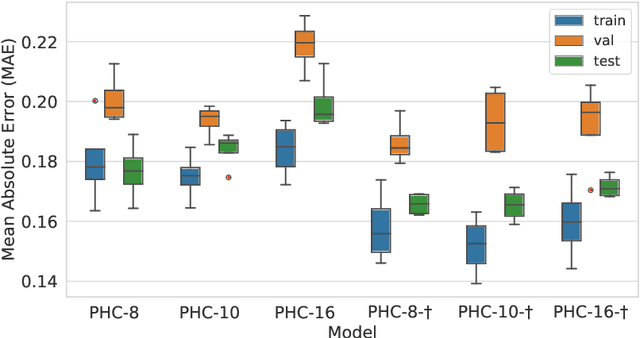
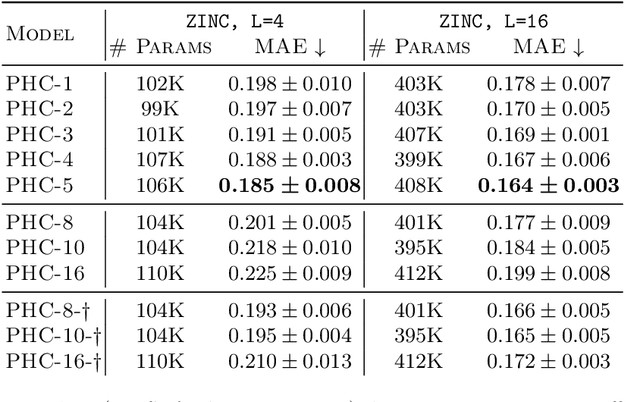
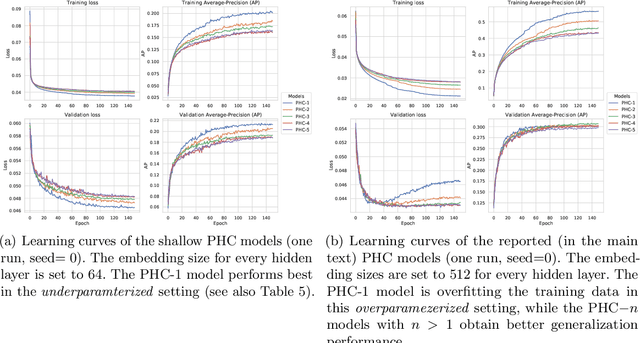
Abstract:Despite recent advances in representation learning in hypercomplex (HC) space, this subject is still vastly unexplored in the context of graphs. Motivated by the complex and quaternion algebras, which have been found in several contexts to enable effective representation learning that inherently incorporates a weight-sharing mechanism, we develop graph neural networks that leverage the properties of hypercomplex feature transformation. In particular, in our proposed class of models, the multiplication rule specifying the algebra itself is inferred from the data during training. Given a fixed model architecture, we present empirical evidence that our proposed model incorporates a regularization effect, alleviating the risk of overfitting. We also show that for fixed model capacity, our proposed method outperforms its corresponding real-formulated GNN, providing additional confirmation for the enhanced expressivity of HC embeddings. Finally, we test our proposed hypercomplex GNN on several open graph benchmark datasets and show that our models reach state-of-the-art performance while consuming a much lower memory footprint with 70& fewer parameters. Our implementations are available at https://github.com/bayer-science-for-a-better-life/phc-gnn.
Autonomous Robotic System using Non-Destructive Evaluation methods for Bridge Deck Inspection
Apr 15, 2017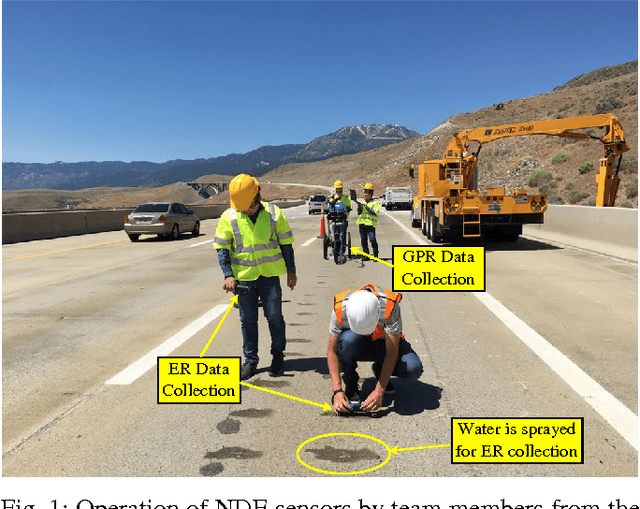
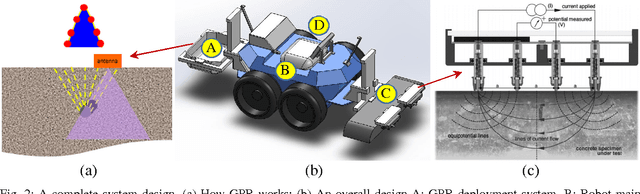
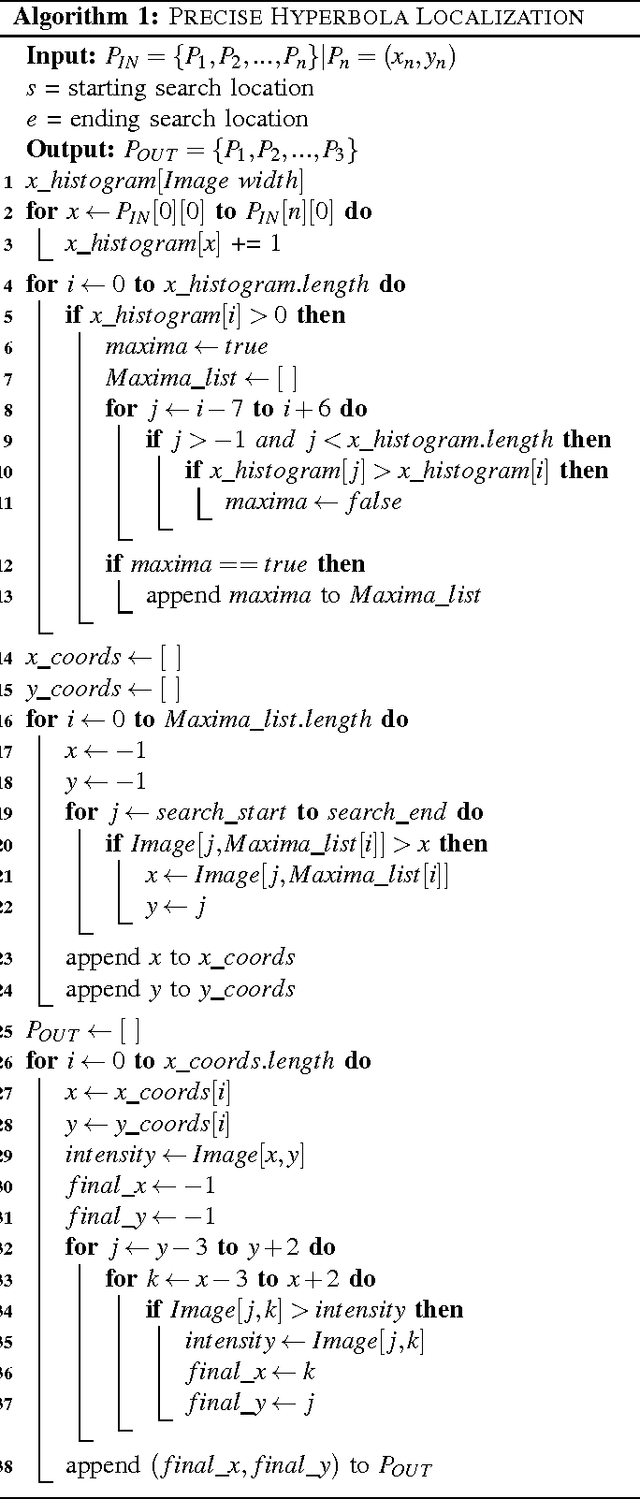
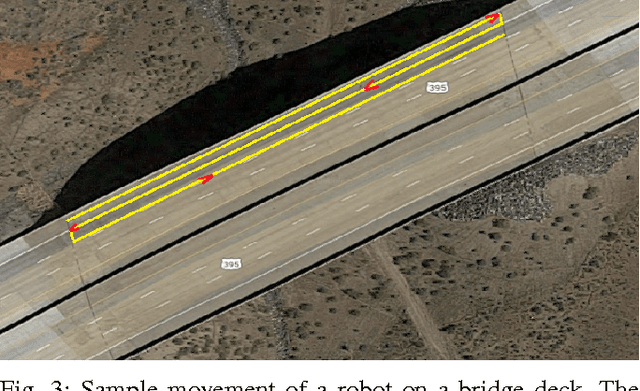
Abstract:Bridge condition assessment is important to maintain the quality of highway roads for public transport. Bridge deterioration with time is inevitable due to aging material, environmental wear and in some cases, inadequate maintenance. Non-destructive evaluation (NDE) methods are preferred for condition assessment for bridges, concrete buildings, and other civil structures. Some examples of NDE methods are ground penetrating radar (GPR), acoustic emission, and electrical resistivity (ER). NDE methods provide the ability to inspect a structure without causing any damage to the structure in the process. In addition, NDE methods typically cost less than other methods, since they do not require inspection sites to be evacuated prior to inspection, which greatly reduces the cost of safety related issues during the inspection process. In this paper, an autonomous robotic system equipped with three different NDE sensors is presented. The system employs GPR, ER, and a camera for data collection. The system is capable of performing real-time, cost-effective bridge deck inspection, and is comprised of a mechanical robot design and machine learning and pattern recognition methods for automated steel rebar picking to provide realtime condition maps of the corrosive deck environments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge