Tuan Lai
Translation between Molecules and Natural Language
Apr 26, 2022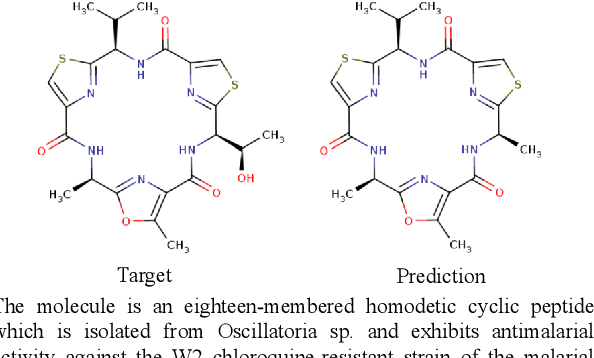
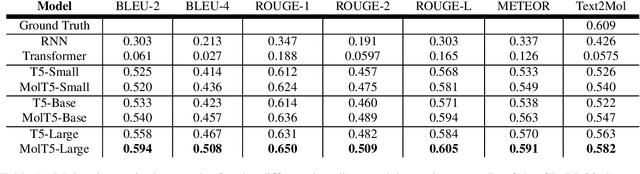
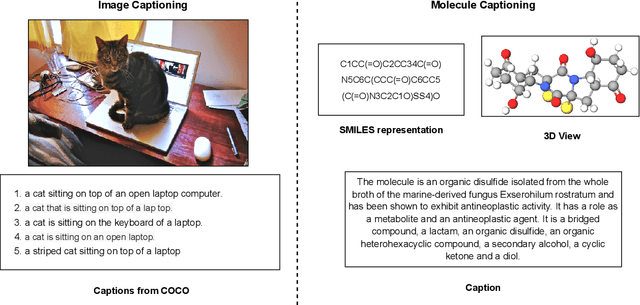
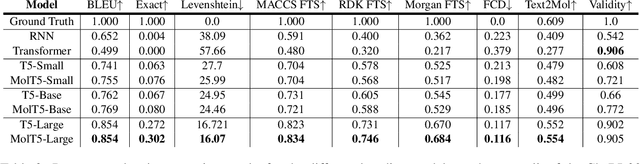
Abstract:Joint representations between images and text have been deeply investigated in the literature. In computer vision, the benefits of incorporating natural language have become clear for enabling semantic-level control of images. In this work, we present $\textbf{MolT5}-$a self-supervised learning framework for pretraining models on a vast amount of unlabeled natural language text and molecule strings. $\textbf{MolT5}$ allows for new, useful, and challenging analogs of traditional vision-language tasks, such as molecule captioning and text-based de novo molecule generation (altogether: translation between molecules and language), which we explore for the first time. Furthermore, since $\textbf{MolT5}$ pretrains models on single-modal data, it helps overcome the chemistry domain shortcoming of data scarcity. Additionally, we consider several metrics, including a new cross-modal embedding-based metric, to evaluate the tasks of molecule captioning and text-based molecule generation. By interfacing molecules with natural language, we enable a higher semantic level of control over molecule discovery and understanding--a critical task for scientific domains such as drug discovery and material design. Our results show that $\textbf{MolT5}$-based models are able to generate outputs, both molecule and text, which in many cases are high quality and match the input modality. On molecule generation, our best model achieves 30% exact matching test accuracy (i.e., it generates the correct structure for about one-third of the captions in our held-out test set).
BERT might be Overkill: A Tiny but Effective Biomedical Entity Linker based on Residual Convolutional Neural Networks
Sep 06, 2021



Abstract:Biomedical entity linking is the task of linking entity mentions in a biomedical document to referent entities in a knowledge base. Recently, many BERT-based models have been introduced for the task. While these models have achieved competitive results on many datasets, they are computationally expensive and contain about 110M parameters. Little is known about the factors contributing to their impressive performance and whether the over-parameterization is needed. In this work, we shed some light on the inner working mechanisms of these large BERT-based models. Through a set of probing experiments, we have found that the entity linking performance only changes slightly when the input word order is shuffled or when the attention scope is limited to a fixed window size. From these observations, we propose an efficient convolutional neural network with residual connections for biomedical entity linking. Because of the sparse connectivity and weight sharing properties, our model has a small number of parameters and is highly efficient. On five public datasets, our model achieves comparable or even better linking accuracy than the state-of-the-art BERT-based models while having about 60 times fewer parameters.
Joint Biomedical Entity and Relation Extraction with Knowledge-Enhanced Collective Inference
Jun 01, 2021



Abstract:Compared to the general news domain, information extraction (IE) from biomedical text requires much broader domain knowledge. However, many previous IE methods do not utilize any external knowledge during inference. Due to the exponential growth of biomedical publications, models that do not go beyond their fixed set of parameters will likely fall behind. Inspired by how humans look up relevant information to comprehend a scientific text, we present a novel framework that utilizes external knowledge for joint entity and relation extraction named KECI (Knowledge-Enhanced Collective Inference). Given an input text, KECI first constructs an initial span graph representing its initial understanding of the text. It then uses an entity linker to form a knowledge graph containing relevant background knowledge for the the entity mentions in the text. To make the final predictions, KECI fuses the initial span graph and the knowledge graph into a more refined graph using an attention mechanism. KECI takes a collective approach to link mention spans to entities by integrating global relational information into local representations using graph convolutional networks. Our experimental results show that the framework is highly effective, achieving new state-of-the-art results in two different benchmark datasets: BioRelEx (binding interaction detection) and ADE (adverse drug event extraction). For example, KECI achieves absolute improvements of 4.59% and 4.91% in F1 scores over the state-of-the-art on the BioRelEx entity and relation extraction tasks.
A Context-Dependent Gated Module for Incorporating Symbolic Semantics into Event Coreference Resolution
Apr 04, 2021



Abstract:Event coreference resolution is an important research problem with many applications. Despite the recent remarkable success of pretrained language models, we argue that it is still highly beneficial to utilize symbolic features for the task. However, as the input for coreference resolution typically comes from upstream components in the information extraction pipeline, the automatically extracted symbolic features can be noisy and contain errors. Also, depending on the specific context, some features can be more informative than others. Motivated by these observations, we propose a novel context-dependent gated module to adaptively control the information flows from the input symbolic features. Combined with a simple noisy training method, our best models achieve state-of-the-art results on two datasets: ACE 2005 and KBP 2016.
AutoNLU: An On-demand Cloud-based Natural Language Understanding System for Enterprises
Nov 26, 2020



Abstract:With the renaissance of deep learning, neural networks have achieved promising results on many natural language understanding (NLU) tasks. Even though the source codes of many neural network models are publicly available, there is still a large gap from open-sourced models to solving real-world problems in enterprises. Therefore, to fill this gap, we introduce AutoNLU, an on-demand cloud-based system with an easy-to-use interface that covers all common use-cases and steps in developing an NLU model. AutoNLU has supported many product teams within Adobe with different use-cases and datasets, quickly delivering them working models. To demonstrate the effectiveness of AutoNLU, we present two case studies. i) We build a practical NLU model for handling various image-editing requests in Photoshop. ii) We build powerful keyphrase extraction models that achieve state-of-the-art results on two public benchmarks. In both cases, end users only need to write a small amount of code to convert their datasets into a common format used by AutoNLU.
Explain by Evidence: An Explainable Memory-based Neural Network for Question Answering
Nov 05, 2020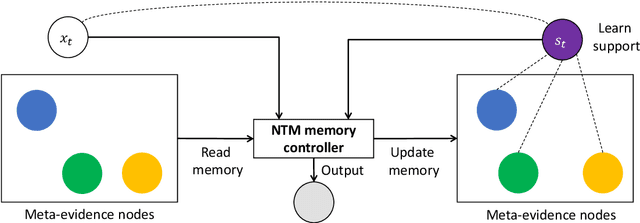
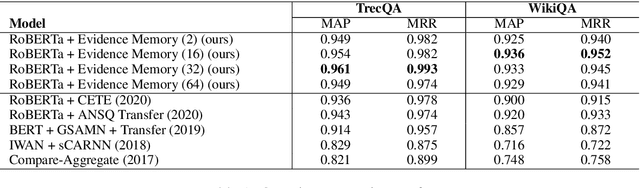
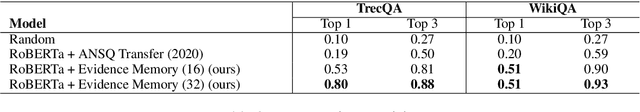
Abstract:Interpretability and explainability of deep neural networks are challenging due to their scale, complexity, and the agreeable notions on which the explaining process rests. Previous work, in particular, has focused on representing internal components of neural networks through human-friendly visuals and concepts. On the other hand, in real life, when making a decision, human tends to rely on similar situations and/or associations in the past. Hence arguably, a promising approach to make the model transparent is to design it in a way such that the model explicitly connects the current sample with the seen ones, and bases its decision on these samples. Grounded on that principle, we propose in this paper an explainable, evidence-based memory network architecture, which learns to summarize the dataset and extract supporting evidences to make its decision. Our model achieves state-of-the-art performance on two popular question answering datasets (i.e. TrecQA and WikiQA). Via further analysis, we show that this model can reliably trace the errors it has made in the validation step to the training instances that might have caused these errors. We believe that this error-tracing capability provides significant benefit in improving dataset quality in many applications.
A Gated Self-attention Memory Network for Answer Selection
Sep 13, 2019


Abstract:Answer selection is an important research problem, with applications in many areas. Previous deep learning based approaches for the task mainly adopt the Compare-Aggregate architecture that performs word-level comparison followed by aggregation. In this work, we take a departure from the popular Compare-Aggregate architecture, and instead, propose a new gated self-attention memory network for the task. Combined with a simple transfer learning technique from a large-scale online corpus, our model outperforms previous methods by a large margin, achieving new state-of-the-art results on two standard answer selection datasets: TrecQA and WikiQA.
A System for Automated Image Editing from Natural Language Commands
Dec 03, 2018



Abstract:This work presents the task of modifying images in an image editing program using natural language written commands. We utilize a corpus of over 6000 image edit text requests to alter real world images collected via crowdsourcing. A novel framework composed of actions and entities to map a user's natural language request to executable commands in an image editing program is described. We resolve previously labeled annotator disagreement through a voting process and complete annotation of the corpus. We experimented with different machine learning models and found that the LSTM, the SVM, and the bidirectional LSTM-CRF joint models are the best to detect image editing actions and associated entities in a given utterance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge