Sophie Merz
On the Value of PHH3 for Mitotic Figure Detection on H&E-stained Images
Jun 28, 2024Abstract:The count of mitotic figures (MFs) observed in hematoxylin and eosin (H&E)-stained slides is an important prognostic marker as it is a measure for tumor cell proliferation. However, the identification of MFs has a known low inter-rater agreement. Deep learning algorithms can standardize this task, but they require large amounts of annotated data for training and validation. Furthermore, label noise introduced during the annotation process may impede the algorithm's performance. Unlike H&E, the mitosis-specific antibody phospho-histone H3 (PHH3) specifically highlights MFs. Counting MFs on slides stained against PHH3 leads to higher agreement among raters and has therefore recently been used as a ground truth for the annotation of MFs in H&E. However, as PHH3 facilitates the recognition of cells indistinguishable from H&E stain alone, the use of this ground truth could potentially introduce noise into the H&E-related dataset, impacting model performance. This study analyzes the impact of PHH3-assisted MF annotation on inter-rater reliability and object level agreement through an extensive multi-rater experiment. We found that the annotators' object-level agreement increased when using PHH3-assisted labeling. Subsequently, MF detectors were evaluated on the resulting datasets to investigate the influence of PHH3-assisted labeling on the models' performance. Additionally, a novel dual-stain MF detector was developed to investigate the interpretation-shift of PHH3-assisted labels used in H&E, which clearly outperformed single-stain detectors. However, the PHH3-assisted labels did not have a positive effect on solely H&E-based models. The high performance of our dual-input detector reveals an information mismatch between the H&E and PHH3-stained images as the cause of this effect.
Nuclear Morphometry using a Deep Learning-based Algorithm has Prognostic Relevance for Canine Cutaneous Mast Cell Tumors
Sep 26, 2023Abstract:Variation in nuclear size and shape is an important criterion of malignancy for many tumor types; however, categorical estimates by pathologists have poor reproducibility. Measurements of nuclear characteristics (morphometry) can improve reproducibility, but manual methods are time consuming. In this study, we evaluated fully automated morphometry using a deep learning-based algorithm in 96 canine cutaneous mast cell tumors with information on patient survival. Algorithmic morphometry was compared with karyomegaly estimates by 11 pathologists, manual nuclear morphometry of 12 cells by 9 pathologists, and the mitotic count as a benchmark. The prognostic value of automated morphometry was high with an area under the ROC curve regarding the tumor-specific survival of 0.943 (95% CI: 0.889 - 0.996) for the standard deviation (SD) of nuclear area, which was higher than manual morphometry of all pathologists combined (0.868, 95% CI: 0.737 - 0.991) and the mitotic count (0.885, 95% CI: 0.765 - 1.00). At the proposed thresholds, the hazard ratio for algorithmic morphometry (SD of nuclear area $\geq 9.0 \mu m^2$) was 18.3 (95% CI: 5.0 - 67.1), for manual morphometry (SD of nuclear area $\geq 10.9 \mu m^2$) 9.0 (95% CI: 6.0 - 13.4), for karyomegaly estimates 7.6 (95% CI: 5.7 - 10.1), and for the mitotic count 30.5 (95% CI: 7.8 - 118.0). Inter-rater reproducibility for karyomegaly estimates was fair ($\kappa$ = 0.226) with highly variable sensitivity/specificity values for the individual pathologists. Reproducibility for manual morphometry (SD of nuclear area) was good (ICC = 0.654). This study supports the use of algorithmic morphometry as a prognostic test to overcome the limitations of estimates and manual measurements.
How Many Annotators Do We Need? -- A Study on the Influence of Inter-Observer Variability on the Reliability of Automatic Mitotic Figure Assessment
Jan 08, 2021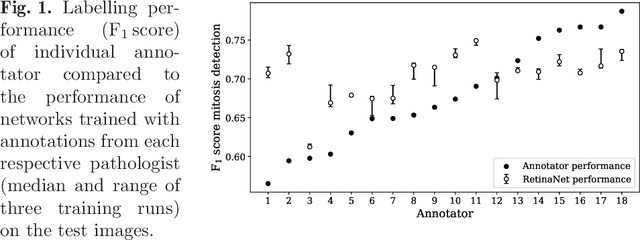
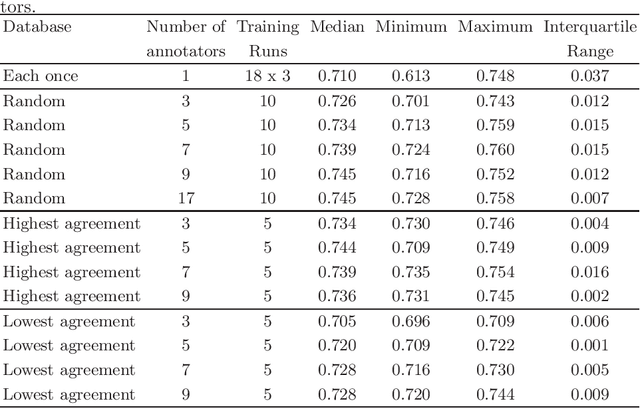
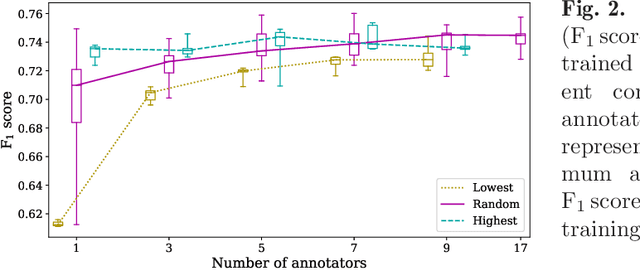
Abstract:Density of mitotic figures in histologic sections is a prognostically relevant characteristic for many tumours. Due to high inter-pathologist variability, deep learning-based algorithms are a promising solution to improve tumour prognostication. Pathologists are the gold standard for database development, however, labelling errors may hamper development of accurate algorithms. In the present work we evaluated the benefit of multi-expert consensus (n = 3, 5, 7, 9, 11) on algorithmic performance. While training with individual databases resulted in highly variable F$_1$ scores, performance was notably increased and more consistent when using the consensus of three annotators. Adding more annotators only resulted in minor improvements. We conclude that databases by few pathologists and high label accuracy may be the best compromise between high algorithmic performance and time investment.
Field of Interest Prediction for Computer-Aided Mitotic Count
Feb 12, 2019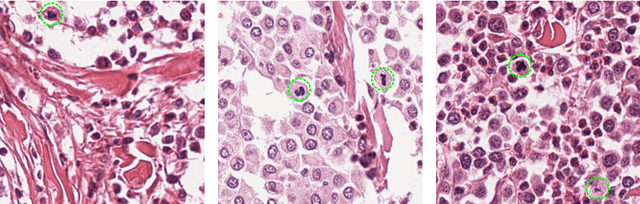
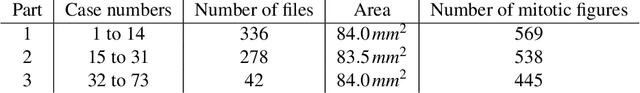
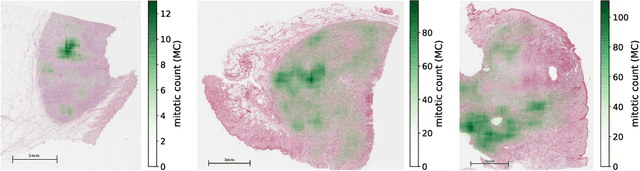
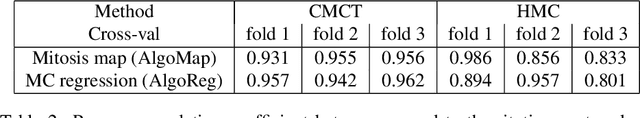
Abstract:Manual counts of mitotic figures, which are determined in the tumor region with the highest mitotic activity, are a key parameter of most tumor grading schemes. It is however strongly dependent on the area selection. To reduce potential variability of prognosis due to this, we propose to use an algorithmic field of interest prediction to assess the area of highest mitotic activity in a whole-slide image. Methods: We evaluated two state-of-the-art methods, all based on the use of deep convolutional neural networks on their ability to predict the mitotic count in digital histopathology slides. We evaluated them on a novel dataset of 32 completely annotated whole slide images from canine cutaneous mast cell tumors (CMCT) and one publicly available human mamma carcinoma (HMC) dataset. We first compared the mitotic counts (MC) predicted by the two models with the ground truth MC on both data sets. Second, for the CMCT data set, we compared the computationally predicted position and MC of the area of highest mitotic activity with size-equivalent areas selected by eight veterinary pathologists. Results: We found a high correlation between the mitotic count as predicted by the models (Pearson's correlation coefficient between 0.931 and 0.962 for the CMCT data set and between 0.801 and 0.986 for the HMC data set) on the slides. For the CMCT data set, this is also reflected in the predicted position representing mitotic counts in mostly the upper quartile of the slide's ground truth MC distribution. Further, we found strong differences between experts in position selection. Conclusion: While the mitotic counts in areas selected by the experts substantially varied, both algorithmic approaches were consistently able to generate a good estimate of the area of highest mitotic count. To achieve better inter-rater agreement, we propose to use computer-based area selection for manual mitotic count.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge