Field of Interest Prediction for Computer-Aided Mitotic Count
Paper and Code
Feb 12, 2019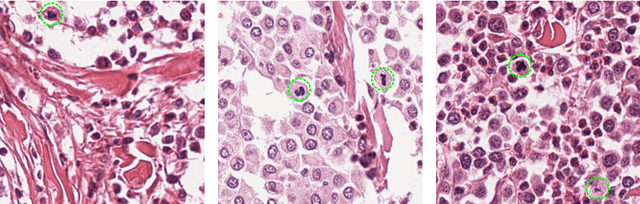
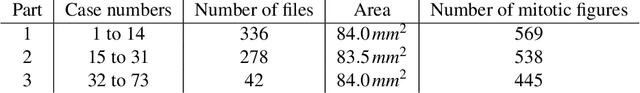
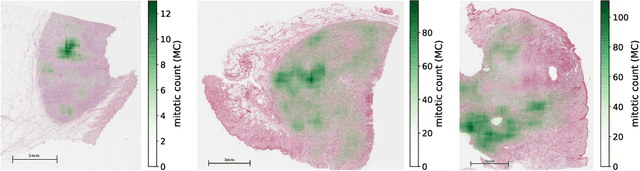
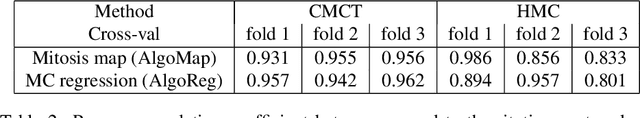
Manual counts of mitotic figures, which are determined in the tumor region with the highest mitotic activity, are a key parameter of most tumor grading schemes. It is however strongly dependent on the area selection. To reduce potential variability of prognosis due to this, we propose to use an algorithmic field of interest prediction to assess the area of highest mitotic activity in a whole-slide image. Methods: We evaluated two state-of-the-art methods, all based on the use of deep convolutional neural networks on their ability to predict the mitotic count in digital histopathology slides. We evaluated them on a novel dataset of 32 completely annotated whole slide images from canine cutaneous mast cell tumors (CMCT) and one publicly available human mamma carcinoma (HMC) dataset. We first compared the mitotic counts (MC) predicted by the two models with the ground truth MC on both data sets. Second, for the CMCT data set, we compared the computationally predicted position and MC of the area of highest mitotic activity with size-equivalent areas selected by eight veterinary pathologists. Results: We found a high correlation between the mitotic count as predicted by the models (Pearson's correlation coefficient between 0.931 and 0.962 for the CMCT data set and between 0.801 and 0.986 for the HMC data set) on the slides. For the CMCT data set, this is also reflected in the predicted position representing mitotic counts in mostly the upper quartile of the slide's ground truth MC distribution. Further, we found strong differences between experts in position selection. Conclusion: While the mitotic counts in areas selected by the experts substantially varied, both algorithmic approaches were consistently able to generate a good estimate of the area of highest mitotic count. To achieve better inter-rater agreement, we propose to use computer-based area selection for manual mitotic count.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge