Shouhei Hanaoka
The Department of Radiology, The University of Tokyo Hospital
Automated Classification of Normal and Atypical Mitotic Figures Using ConvNeXt V2: MIDOG 2025 Track 2
Aug 26, 2025Abstract:This paper presents our solution for the MIDOG 2025 Challenge Track 2, which focuses on binary classification of normal mitotic figures (NMFs) versus atypical mitotic figures (AMFs) in histopathological images. Our approach leverages a ConvNeXt V2 base model with center cropping preprocessing and 5-fold cross-validation ensemble strategy. The method addresses key challenges including severe class imbalance, high morphological variability, and domain heterogeneity across different tumor types, species, and scanners. Through strategic preprocessing with 60% center cropping and mixed precision training, our model achieved robust performance on the diverse MIDOG 2025 dataset. The solution demonstrates the effectiveness of modern convolutional architectures for mitotic figure subtyping while maintaining computational efficiency through careful architectural choices and training optimizations.
CXR-LT 2024: A MICCAI challenge on long-tailed, multi-label, and zero-shot disease classification from chest X-ray
Jun 09, 2025Abstract:The CXR-LT series is a community-driven initiative designed to enhance lung disease classification using chest X-rays (CXR). It tackles challenges in open long-tailed lung disease classification and enhances the measurability of state-of-the-art techniques. The first event, CXR-LT 2023, aimed to achieve these goals by providing high-quality benchmark CXR data for model development and conducting comprehensive evaluations to identify ongoing issues impacting lung disease classification performance. Building on the success of CXR-LT 2023, the CXR-LT 2024 expands the dataset to 377,110 chest X-rays (CXRs) and 45 disease labels, including 19 new rare disease findings. It also introduces a new focus on zero-shot learning to address limitations identified in the previous event. Specifically, CXR-LT 2024 features three tasks: (i) long-tailed classification on a large, noisy test set, (ii) long-tailed classification on a manually annotated "gold standard" subset, and (iii) zero-shot generalization to five previously unseen disease findings. This paper provides an overview of CXR-LT 2024, detailing the data curation process and consolidating state-of-the-art solutions, including the use of multimodal models for rare disease detection, advanced generative approaches to handle noisy labels, and zero-shot learning strategies for unseen diseases. Additionally, the expanded dataset enhances disease coverage to better represent real-world clinical settings, offering a valuable resource for future research. By synthesizing the insights and innovations of participating teams, we aim to advance the development of clinically realistic and generalizable diagnostic models for chest radiography.
ModernBERT is More Efficient than Conventional BERT for Chest CT Findings Classification in Japanese Radiology Reports
Mar 07, 2025



Abstract:Objective: This study aims to evaluate and compare the performance of two Japanese language models-conventional Bidirectional Encoder Representations from Transformers (BERT) and the newer ModernBERT-in classifying findings from chest CT reports, with a focus on tokenization efficiency, processing time, and classification performance. Methods: We conducted a retrospective study using the CT-RATE-JPN dataset containing 22,778 training reports and 150 test reports. Both models were fine-tuned for multi-label classification of 18 common chest CT conditions. The training data was split in 18,222:4,556 for training and validation. Performance was evaluated using F1 scores for each condition and exact match accuracy across all 18 labels. Results: ModernBERT demonstrated superior tokenization efficiency, requiring 24.0% fewer tokens per document (258.1 vs. 339.6) compared to BERT Base. This translated to significant performance improvements, with ModernBERT completing training in 1877.67 seconds versus BERT's 3090.54 seconds (39% reduction). ModernBERT processed 38.82 samples per second during training (1.65x faster) and 139.90 samples per second during inference (1.66x faster). Despite these efficiency gains, classification performance remained comparable, with ModernBERT achieving superior F1 scores in 8 conditions, while BERT performed better in 4 conditions. Overall exact match accuracy was slightly higher for ModernBERT (74.67% vs. 72.67%), though this difference was not statistically significant (p=0.6291). Conclusion: ModernBERT offers substantial improvements in tokenization efficiency and training speed without sacrificing classification performance. These results suggest that ModernBERT is a promising candidate for clinical applications in Japanese radiology reports analysis.
Development of a Large-scale Dataset of Chest Computed Tomography Reports in Japanese and a High-performance Finding Classification Model
Dec 20, 2024



Abstract:Background: Recent advances in large language models highlight the need for high-quality multilingual medical datasets. While Japan leads globally in CT scanner deployment and utilization, the lack of large-scale Japanese radiology datasets has hindered the development of specialized language models for medical imaging analysis. Objective: To develop a comprehensive Japanese CT report dataset through machine translation and establish a specialized language model for structured finding classification. Additionally, to create a rigorously validated evaluation dataset through expert radiologist review. Methods: We translated the CT-RATE dataset (24,283 CT reports from 21,304 patients) into Japanese using GPT-4o mini. The training dataset consisted of 22,778 machine-translated reports, while the validation dataset included 150 radiologist-revised reports. We developed CT-BERT-JPN based on "tohoku-nlp/bert-base-japanese-v3" architecture for extracting 18 structured findings from Japanese radiology reports. Results: Translation metrics showed strong performance with BLEU scores of 0.731 and 0.690, and ROUGE scores ranging from 0.770 to 0.876 for Findings and from 0.748 to 0.857 for Impression sections. CT-BERT-JPN demonstrated superior performance compared to GPT-4o in 11 out of 18 conditions, including lymphadenopathy (+14.2%), interlobular septal thickening (+10.9%), and atelectasis (+7.4%). The model maintained F1 scores exceeding 0.95 in 14 out of 18 conditions and achieved perfect scores in four conditions. Conclusions: Our study establishes a robust Japanese CT report dataset and demonstrates the effectiveness of a specialized language model for structured finding classification. The hybrid approach of machine translation and expert validation enables the creation of large-scale medical datasets while maintaining high quality.
Ensemble of ConvNeXt V2 and MaxViT for Long-Tailed CXR Classification with View-Based Aggregation
Oct 15, 2024

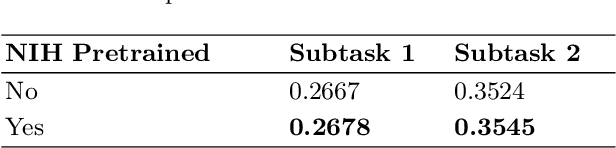
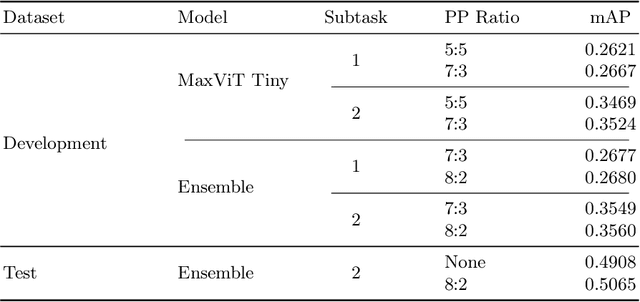
Abstract:In this work, we present our solution for the MICCAI 2024 CXR-LT challenge, achieving 4th place in Subtask 2 and 5th in Subtask 1. We leveraged an ensemble of ConvNeXt V2 and MaxViT models, pretrained on an external chest X-ray dataset, to address the long-tailed distribution of chest findings. The proposed method combines state-of-the-art image classification techniques, asymmetric loss for handling class imbalance, and view-based prediction aggregation to enhance classification performance. Through experiments, we demonstrate the advantages of our approach in improving both detection accuracy and the handling of the long-tailed distribution in CXR findings. The code is available at https://github.com/yamagishi0824/cxrlt24-multiview-pp.
Zero-shot 3D Segmentation of Abdominal Organs in CT Scans Using Segment Anything Model 2: Adapting Video Tracking Capabilities for 3D Medical Imaging
Aug 12, 2024

Abstract:Purpose: This study aimed to evaluate the zero-shot performance of Segment Anything Model 2 (SAM 2) in 3D segmentation of abdominal organs in CT scans, leveraging its video tracking capabilities for volumetric medical imaging. Materials and Methods: Using a subset of the TotalSegmentator CT dataset (n=123) from 8 different institutions, we assessed SAM 2's ability to segment 8 abdominal organs. Segmentation was initiated from three different Z-coordinate levels (caudal, mid, and cranial levels) of each organ. Performance was measured using the Dice similarity coefficient (DSC). We also analyzed organ volumes to contextualize the results. Results: As a zero-shot approach, larger organs with clear boundaries demonstrated high segmentation performance, with mean(median) DSCs as follows: liver 0.821(0.898), left kidney 0.870(0.921), right kidney 0.862(0.935), and spleen 0.891(0.932). Smaller or less defined structures showed lower performance: gallbladder 0.531(0.590), pancreas 0.361(0.359), and adrenal glands 0.203-0.308(0.109-0.231). Significant differences in DSC were observed depending on the starting initial slice of segmentation for different organs. A moderate positive correlation was observed between volume size and DSCs (Spearman's rs = 0.731, P <.001 at caudal-level). DSCs exhibited high variability within organs, ranging from near 0 to almost 1.0, indicating substantial inconsistency in segmentation performance between scans. Conclusion: SAM 2 demonstrated promising zero-shot performance in segmenting certain abdominal organs in CT scans, particularly larger organs with clear boundaries. The model's ability to segment previously unseen targets without additional training highlights its potential for cross-domain generalization in medical imaging. However, improvements are needed for smaller and less defined structures.
Data Set Terminology of Artificial Intelligence in Medicine: A Historical Review and Recommendation
Apr 30, 2024Abstract:Medicine and artificial intelligence (AI) engineering represent two distinct fields each with decades of published history. With such history comes a set of terminology that has a specific way in which it is applied. However, when two distinct fields with overlapping terminology start to collaborate, miscommunication and misunderstandings can occur. This narrative review aims to give historical context for these terms, accentuate the importance of clarity when these terms are used in medical AI contexts, and offer solutions to mitigate misunderstandings by readers from either field. Through an examination of historical documents, including articles, writing guidelines, and textbooks, this review traces the divergent evolution of terms for data sets and their impact. Initially, the discordant interpretations of the word 'validation' in medical and AI contexts are explored. Then the data sets used for AI evaluation are classified, namely random splitting, cross-validation, temporal, geographic, internal, and external sets. The accurate and standardized description of these data sets is crucial for demonstrating the robustness and generalizability of AI applications in medicine. This review clarifies existing literature to provide a comprehensive understanding of these classifications and their implications in AI evaluation. This review then identifies often misunderstood terms and proposes pragmatic solutions to mitigate terminological confusion. Among these solutions are the use of standardized terminology such as 'training set,' 'validation (or tuning) set,' and 'test set,' and explicit definition of data set splitting terminologies in each medical AI research publication. This review aspires to enhance the precision of communication in medical AI, thereby fostering more effective and transparent research methodologies in this interdisciplinary field.
Synthetic data generation method for hybrid image-tabular data using two generative adversarial networks
Aug 15, 2023



Abstract:The generation of synthetic medical records using generative adversarial networks (GANs) has become increasingly important for addressing privacy concerns and promoting data sharing in the medical field. In this paper, we propose a novel method for generating synthetic hybrid medical records consisting of chest X-ray images (CXRs) and structured tabular data (including anthropometric data and laboratory tests) using an auto-encoding GAN ({\alpha}GAN) and a conditional tabular GAN (CTGAN). Our approach involves training a {\alpha}GAN model on a large public database (pDB) to reduce the dimensionality of CXRs. We then applied the trained encoder of the GAN model to the images in original database (oDB) to obtain the latent vectors. These latent vectors were combined with tabular data in oDB, and these joint data were used to train the CTGAN model. We successfully generated diverse synthetic records of hybrid CXR and tabular data, maintaining correspondence between them. We evaluated this synthetic database (sDB) through visual assessment, distribution of interrecord distances, and classification tasks. Our evaluation results showed that the sDB captured the features of the oDB while maintaining the correspondence between the images and tabular data. Although our approach relies on the availability of a large-scale pDB containing a substantial number of images with the same modality and imaging region as those in the oDB, this method has the potential for the public release of synthetic datasets without compromising the secondary use of data.
Local Differential Privacy Image Generation Using Flow-based Deep Generative Models
Dec 20, 2022



Abstract:Diagnostic radiologists need artificial intelligence (AI) for medical imaging, but access to medical images required for training in AI has become increasingly restrictive. To release and use medical images, we need an algorithm that can simultaneously protect privacy and preserve pathologies in medical images. To develop such an algorithm, here, we propose DP-GLOW, a hybrid of a local differential privacy (LDP) algorithm and one of the flow-based deep generative models (GLOW). By applying a GLOW model, we disentangle the pixelwise correlation of images, which makes it difficult to protect privacy with straightforward LDP algorithms for images. Specifically, we map images onto the latent vector of the GLOW model, each element of which follows an independent normal distribution, and we apply the Laplace mechanism to the latent vector. Moreover, we applied DP-GLOW to chest X-ray images to generate LDP images while preserving pathologies.
Aging prediction using deep generative model toward the development of preventive medicine
Aug 23, 2022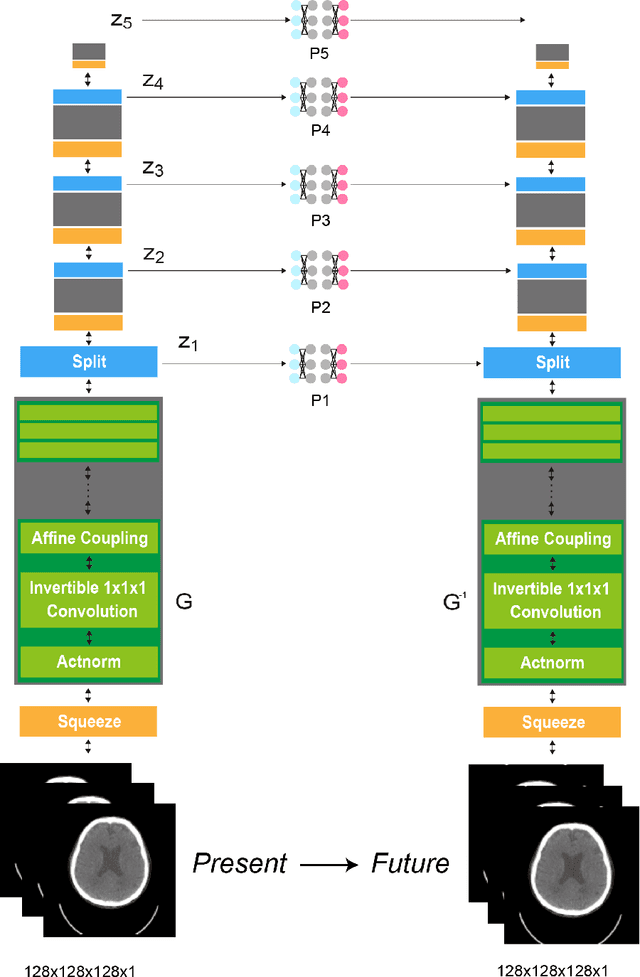

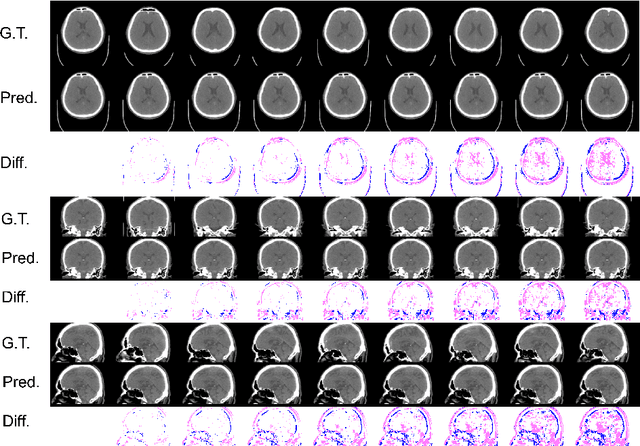
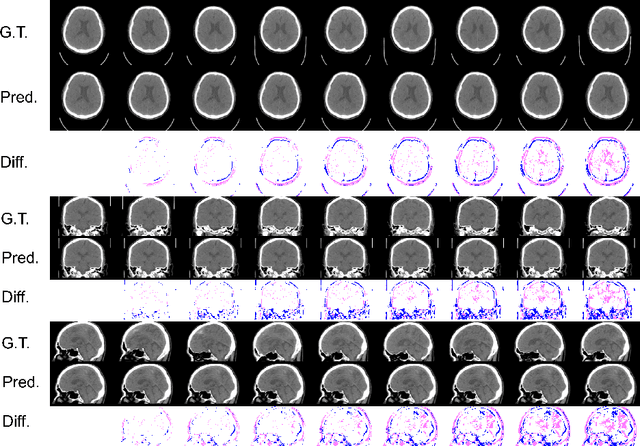
Abstract:From birth to death, we all experience surprisingly ubiquitous changes over time due to aging. If we can predict aging in the digital domain, that is, the digital twin of the human body, we would be able to detect lesions in their very early stages, thereby enhancing the quality of life and extending the life span. We observed that none of the previously developed digital twins of the adult human body explicitly trained longitudinal conversion rules between volumetric medical images with deep generative models, potentially resulting in poor prediction performance of, for example, ventricular volumes. Here, we establish a new digital twin of an adult human body that adopts longitudinally acquired head computed tomography (CT) images for training, enabling prediction of future volumetric head CT images from a single present volumetric head CT image. We, for the first time, adopt one of the three-dimensional flow-based deep generative models to realize this sequential three-dimensional digital twin. We show that our digital twin outperforms the latest methods of prediction of ventricular volumes in relatively short terms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge