Hiroshi Seki
Data Set Terminology of Artificial Intelligence in Medicine: A Historical Review and Recommendation
Apr 30, 2024Abstract:Medicine and artificial intelligence (AI) engineering represent two distinct fields each with decades of published history. With such history comes a set of terminology that has a specific way in which it is applied. However, when two distinct fields with overlapping terminology start to collaborate, miscommunication and misunderstandings can occur. This narrative review aims to give historical context for these terms, accentuate the importance of clarity when these terms are used in medical AI contexts, and offer solutions to mitigate misunderstandings by readers from either field. Through an examination of historical documents, including articles, writing guidelines, and textbooks, this review traces the divergent evolution of terms for data sets and their impact. Initially, the discordant interpretations of the word 'validation' in medical and AI contexts are explored. Then the data sets used for AI evaluation are classified, namely random splitting, cross-validation, temporal, geographic, internal, and external sets. The accurate and standardized description of these data sets is crucial for demonstrating the robustness and generalizability of AI applications in medicine. This review clarifies existing literature to provide a comprehensive understanding of these classifications and their implications in AI evaluation. This review then identifies often misunderstood terms and proposes pragmatic solutions to mitigate terminological confusion. Among these solutions are the use of standardized terminology such as 'training set,' 'validation (or tuning) set,' and 'test set,' and explicit definition of data set splitting terminologies in each medical AI research publication. This review aspires to enhance the precision of communication in medical AI, thereby fostering more effective and transparent research methodologies in this interdisciplinary field.
Reduced-Lead ECG Classifier Model Trained with DivideMix and Model Ensemble
Sep 24, 2021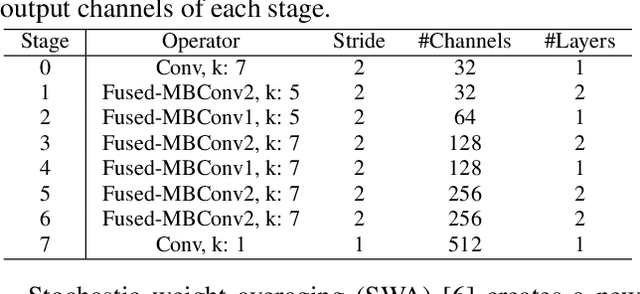
Abstract:Automatic diagnosis of multiple cardiac abnormalities from reduced-lead electrocardiogram (ECG) data is challenging. One of the reasons for this is the difficulty of defining labels from standard 12-lead data. Reduced-lead ECG data usually do not have identical characteristics of cardiac abnormalities because of the noisy label problem. Thus, there is an inconsistency in the annotated labels between the reduced-lead and 12-lead ECG data. To solve this, we propose deep neural network (DNN)-based ECG classifier models that incorporate DivideMix and stochastic weight averaging (SWA). DivideMix was used to refine the noisy label by using two separate models. Besides DivideMix, we used a model ensemble technique, SWA, which also focuses on the noisy label problem, to enhance the effect of the models generated by DivideMix. Our classifiers (ami_kagoshima) received scores of 0.49, 0.47, 0.48, 0.47, and 0.47 (ranked 9th, 10th, 10th, 11th, and 10th, respectively, out of 39 teams) for the 12-lead, 6-lead, 4-lead, 3-lead, and 2-lead versions, respectively, of the hidden test set with the challenge evaluation metric. We obtained the scores of 0.701, 0.686, 0.693, 0.693, and 0.685 on the 10-fold cross validation, and 0.623, 0.593, 0.606, 0.612, and 0.601 on the hidden validation set for each lead combination.
Vectorization of hypotheses and speech for faster beam search in encoder decoder-based speech recognition
Nov 12, 2018



Abstract:Attention-based encoder decoder network uses a left-to-right beam search algorithm in the inference step. The current beam search expands hypotheses and traverses the expanded hypotheses at the next time step. This traversal is implemented using a for-loop program in general, and it leads to speed down of the recognition process. In this paper, we propose a parallelism technique for beam search, which accelerates the search process by vectorizing multiple hypotheses to eliminate the for-loop program. We also propose a technique to batch multiple speech utterances for off-line recognition use, which reduces the for-loop program with regard to the traverse of multiple utterances. This extension is not trivial during beam search unlike during training due to several pruning and thresholding techniques for efficient decoding. In addition, our method can combine scores of external modules, RNNLM and CTC, in a batch as shallow fusion. We achieved 3.7 x speedup compared with the original beam search algorithm by vectoring hypotheses, and achieved 10.5 x speedup by further changing processing unit to GPU.
A Purely End-to-end System for Multi-speaker Speech Recognition
May 15, 2018



Abstract:Recently, there has been growing interest in multi-speaker speech recognition, where the utterances of multiple speakers are recognized from their mixture. Promising techniques have been proposed for this task, but earlier works have required additional training data such as isolated source signals or senone alignments for effective learning. In this paper, we propose a new sequence-to-sequence framework to directly decode multiple label sequences from a single speech sequence by unifying source separation and speech recognition functions in an end-to-end manner. We further propose a new objective function to improve the contrast between the hidden vectors to avoid generating similar hypotheses. Experimental results show that the model is directly able to learn a mapping from a speech mixture to multiple label sequences, achieving 83.1 % relative improvement compared to a model trained without the proposed objective. Interestingly, the results are comparable to those produced by previous end-to-end works featuring explicit separation and recognition modules.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge