Saurabh Johri
Learning medical triage from clinicians using Deep Q-Learning
Mar 28, 2020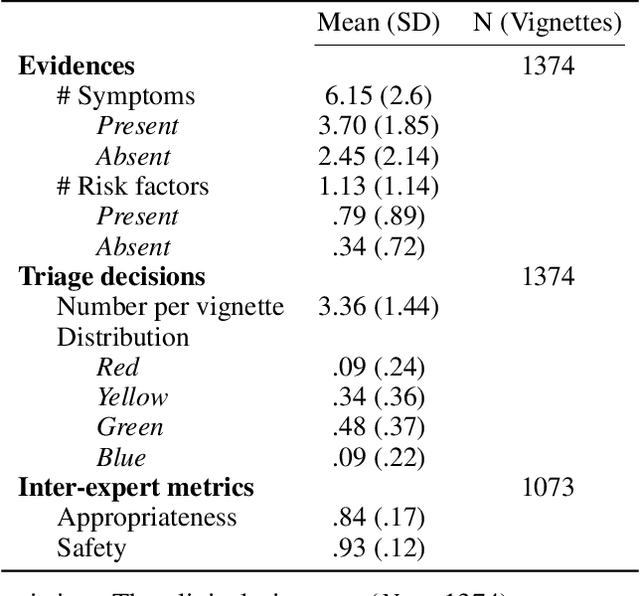
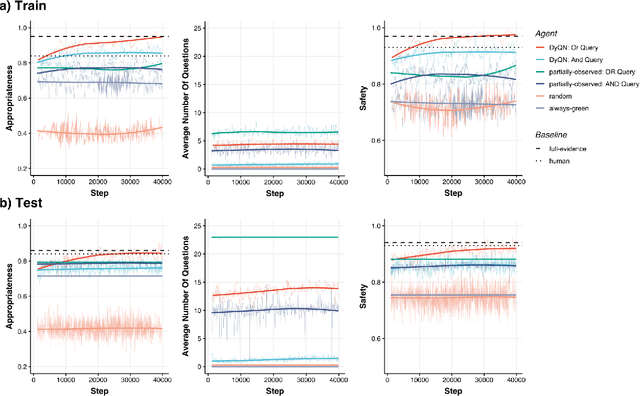
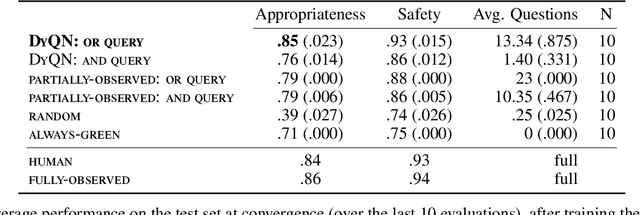
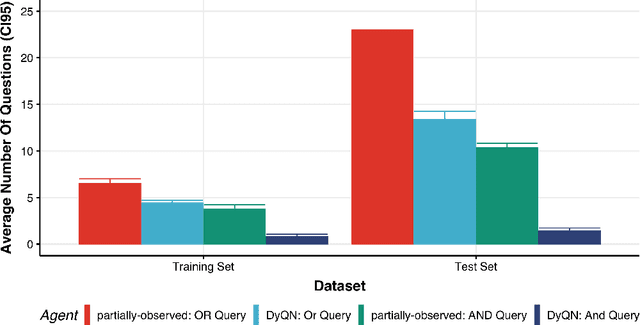
Abstract:Medical Triage is of paramount importance to healthcare systems, allowing for the correct orientation of patients and allocation of the necessary resources to treat them adequately. While reliable decision-tree methods exist to triage patients based on their presentation, those trees implicitly require human inference and are not immediately applicable in a fully automated setting. On the other hand, learning triage policies directly from experts may correct for some of the limitations of hard-coded decision-trees. In this work, we present a Deep Reinforcement Learning approach (a variant of DeepQ-Learning) to triage patients using curated clinical vignettes. The dataset, consisting of 1374 clinical vignettes, was created by medical doctors to represent real-life cases. Each vignette is associated with an average of 3.8 expert triage decisions given by medical doctors relying solely on medical history. We show that this approach is on a par with human performance, yielding safe triage decisions in 94% of cases, and matching expert decisions in 85% of cases. The trained agent learns when to stop asking questions, acquires optimized decision policies requiring less evidence than supervised approaches, and adapts to the novelty of a situation by asking for more information. Overall, we demonstrate that a Deep Reinforcement Learning approach can learn effective medical triage policies directly from expert decisions, without requiring expert knowledge engineering. This approach is scalable and can be deployed in healthcare settings or geographical regions with distinct triage specifications, or where trained experts are scarce, to improve decision making in the early stage of care.
Masking schemes for universal marginalisers
Jan 16, 2020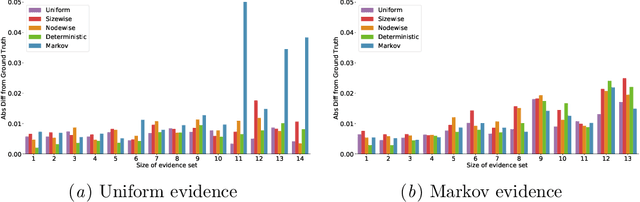
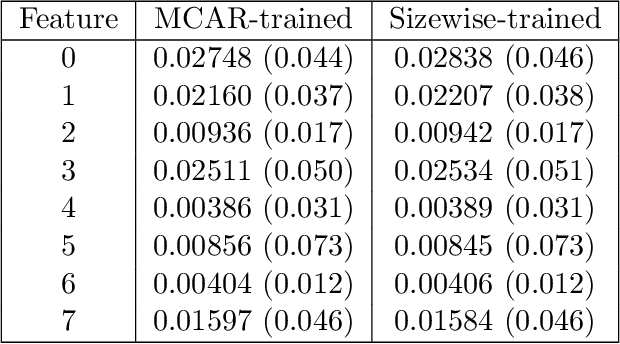

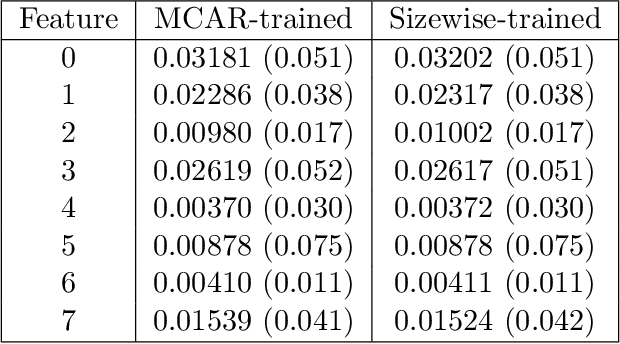
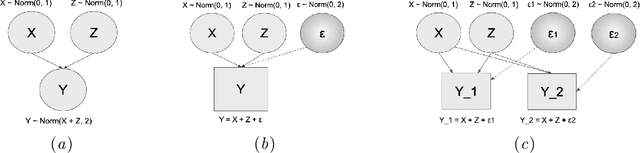
Abstract:We consider the effect of structure-agnostic and structure-dependent masking schemes when training a universal marginaliser (arXiv:1711.00695) in order to learn conditional distributions of the form $P(x_i |\mathbf x_{\mathbf b})$, where $x_i$ is a given random variable and $\mathbf x_{\mathbf b}$ is some arbitrary subset of all random variables of the generative model of interest. In other words, we mimic the self-supervised training of a denoising autoencoder, where a dataset of unlabelled data is used as partially observed input and the neural approximator is optimised to minimise reconstruction loss. We focus on studying the underlying process of the partially observed data---how good is the neural approximator at learning all conditional distributions when the observation process at prediction time differs from the masking process during training? We compare networks trained with different masking schemes in terms of their predictive performance and generalisation properties.
Leveraging directed causal discovery to detect latent common causes
Nov 11, 2019
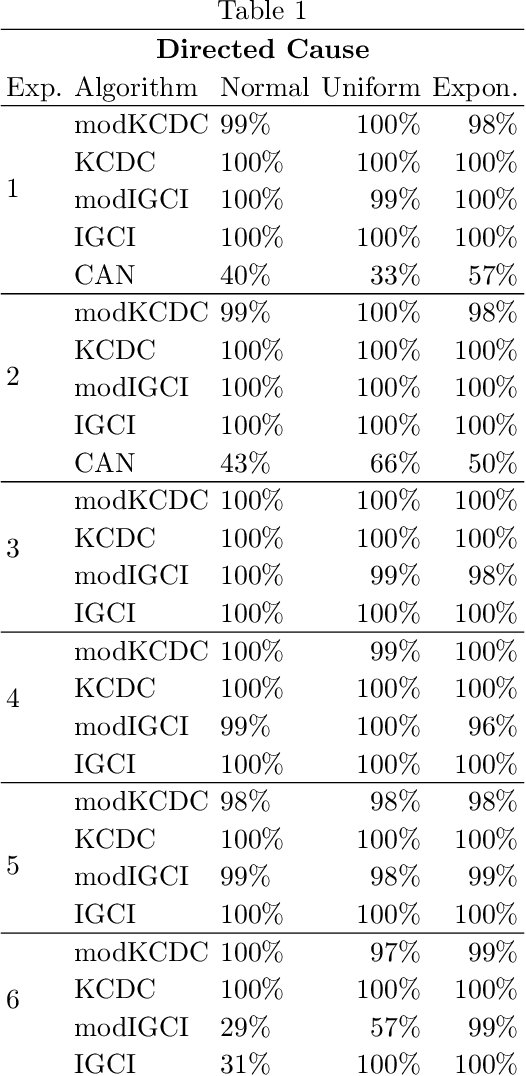
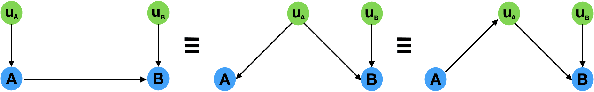

Abstract:The discovery of causal relationships is a fundamental problem in science and medicine. In recent years, many elegant approaches to discovering causal relationships between two variables from uncontrolled data have been proposed. However, most of these deal only with purely directed causal relationships and cannot detect latent common causes. Here, we devise a general method which takes a purely directed causal discovery algorithm and modifies it so that it can also detect latent common causes. The identifiability of the modified algorithm depends on the identifiability of the original, as well as an assumption that the strength of noise be relatively small. We apply our method to two directed causal discovery algorithms, the Information Geometric Causal Inference of (Daniusis et al., 2010) and the Kernel Conditional Deviance for Causal Inference of (Mitrovic, Sejdinovic, and Teh, 2018), and extensively test on synthetic data---detecting latent common causes in additive, multiplicative and complex noise regimes---and on real data, where we are able to detect known common causes. In addition to detecting latent common causes, our experiments demonstrate that both modified algorithms preserve the performance of the original directed algorithm in distinguishing directed causal relations.
Counterfactual diagnosis
Oct 28, 2019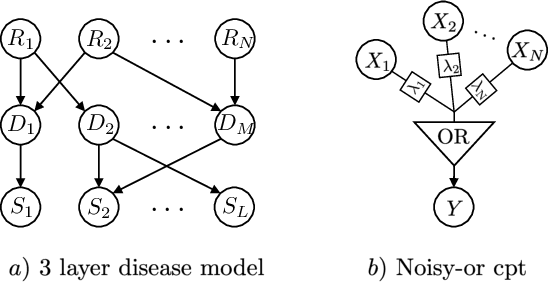
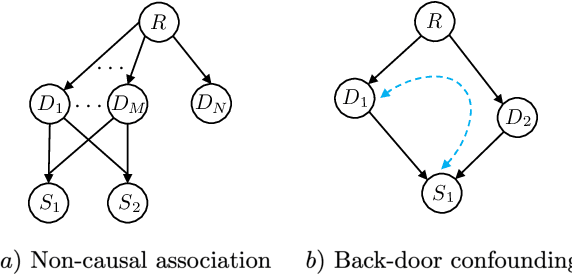
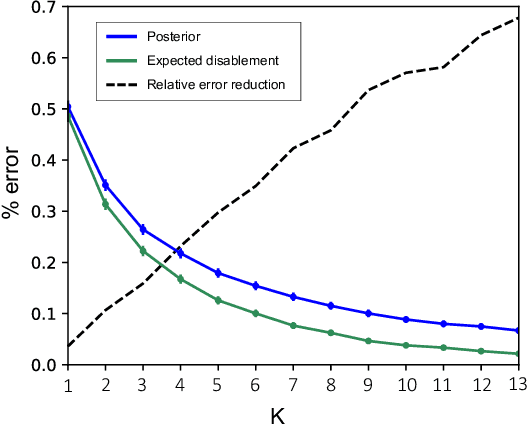
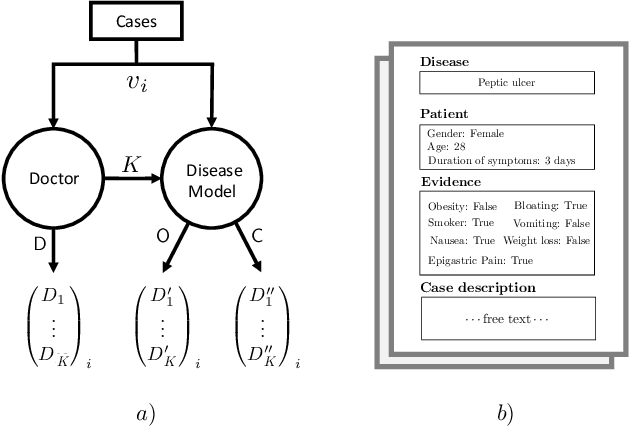
Abstract:Causal knowledge is vital for effective reasoning in science and medicine. In medical diagnosis for example, a doctor aims to explain a patient's symptoms by determining the diseases \emph{causing} them. However, all previous approaches to Machine-Learning assisted diagnosis, including Deep Learning and model-based Bayesian approaches, learn by association and do not distinguish correlation from causation. Here, we propose a new diagnostic algorithm based on counterfactual inference which captures the causal aspect of diagnosis overlooked by previous approaches. Using a statistical disease model, which describes the relations between hundreds of diseases, symptoms and risk factors, we compare our counterfactual algorithm to the standard Bayesian diagnostic algorithm, and test these against a cohort of 44 doctors. We use 1671 clinical vignettes created by a separate panel of doctors to benchmark performance. Each vignette provides a non-exhaustive list of symptoms and medical history simulating a single presentation of a disease. The algorithms and doctors are tasked with determining the underlying disease for each vignette from symptom and medical history information alone. While the Bayesian algorithm achieves an accuracy comparable to the average doctor, placing in the top 48\% of doctors in our cohort, our counterfactual algorithm places in the top 25\% of doctors, achieving expert clinical accuracy. Our results demonstrate the advantage of counterfactual over associative reasoning in a complex real-world task, and show that counterfactual reasoning is a vital missing ingredient for applying machine learning to medical diagnosis.
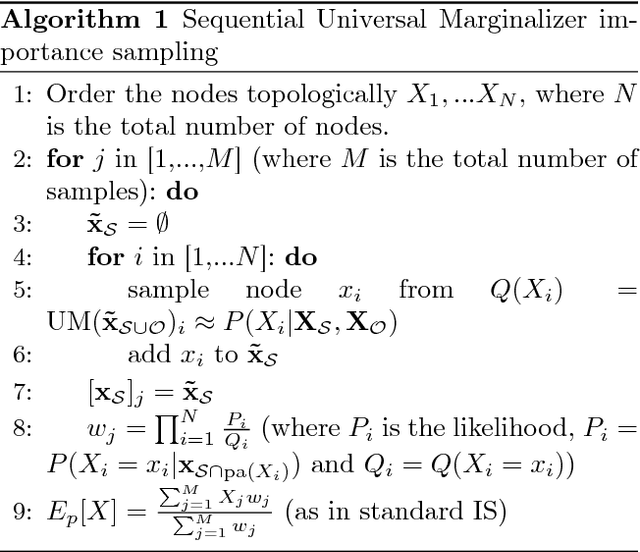
MultiVerse: Causal Reasoning using Importance Sampling in Probabilistic Programming
Oct 17, 2019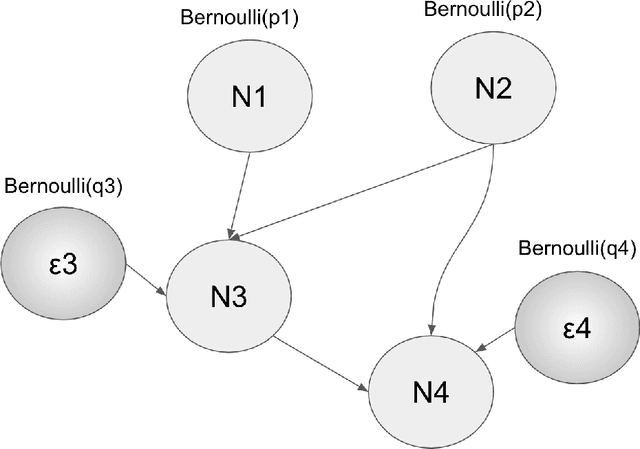
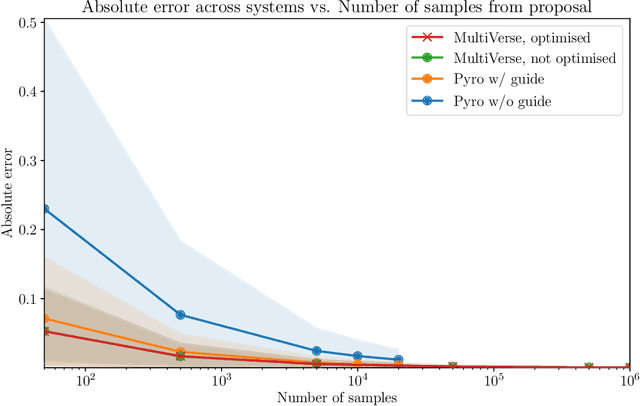
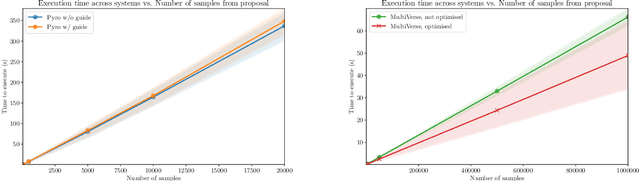
Abstract:We elaborate on using importance sampling for causal reasoning, in particular for counterfactual inference. We show how this can be implemented natively in probabilistic programming. By considering the structure of the counterfactual query, one can significantly optimise the inference process. We also consider design choices to enable further optimisations. We introduce MultiVerse, a probabilistic programming prototype engine for approximate causal reasoning. We provide experimental results and compare with Pyro, an existing probabilistic programming framework with some of causal reasoning tools.
Universal Marginaliser for Deep Amortised Inference for Probabilistic Programs
Oct 16, 2019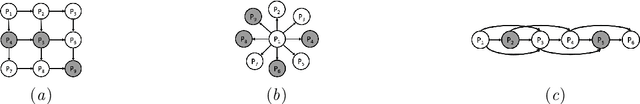
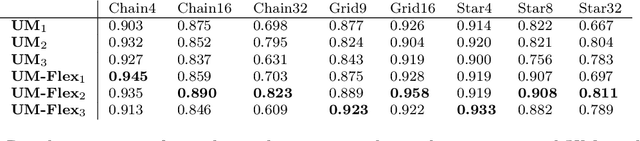
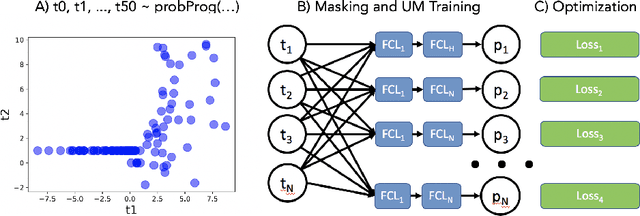
Abstract:Probabilistic programming languages (PPLs) are powerful modelling tools which allow to formalise our knowledge about the world and reason about its inherent uncertainty. Inference methods used in PPL can be computationally costly due to significant time burden and/or storage requirements; or they can lack theoretical guarantees of convergence and accuracy when applied to large scale graphical models. To this end, we present the Universal Marginaliser (UM), a novel method for amortised inference, in PPL. We show how combining samples drawn from the original probabilistic program prior with an appropriate augmentation method allows us to train one neural network to approximate any of the corresponding conditional marginal distributions, with any separation into latent and observed variables, and thus amortise the cost of inference. Finally, we benchmark the method on multiple probabilistic programs, in Pyro, with different model structure.
Universal Marginalizer for Amortised Inference and Embedding of Generative Models
Nov 12, 2018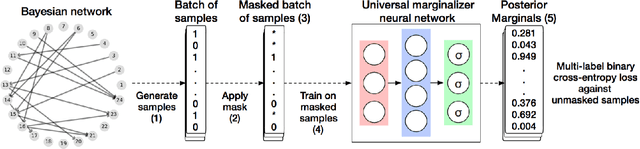
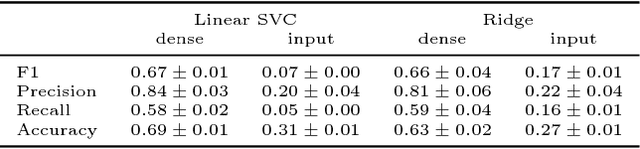

Abstract:Probabilistic graphical models are powerful tools which allow us to formalise our knowledge about the world and reason about its inherent uncertainty. There exist a considerable number of methods for performing inference in probabilistic graphical models; however, they can be computationally costly due to significant time burden and/or storage requirements; or they lack theoretical guarantees of convergence and accuracy when applied to large scale graphical models. To this end, we propose the Universal Marginaliser Importance Sampler (UM-IS) -- a hybrid inference scheme that combines the flexibility of a deep neural network trained on samples from the model and inherits the asymptotic guarantees of importance sampling. We show how combining samples drawn from the graphical model with an appropriate masking function allows us to train a single neural network to approximate any of the corresponding conditional marginal distributions, and thus amortise the cost of inference. We also show that the graph embeddings can be applied for tasks such as: clustering, classification and interpretation of relationships between the nodes. Finally, we benchmark the method on a large graph (>1000 nodes), showing that UM-IS outperforms sampling-based methods by a large margin while being computationally efficient.
A comparative study of artificial intelligence and human doctors for the purpose of triage and diagnosis
Jun 27, 2018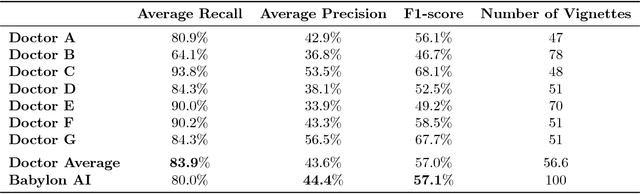
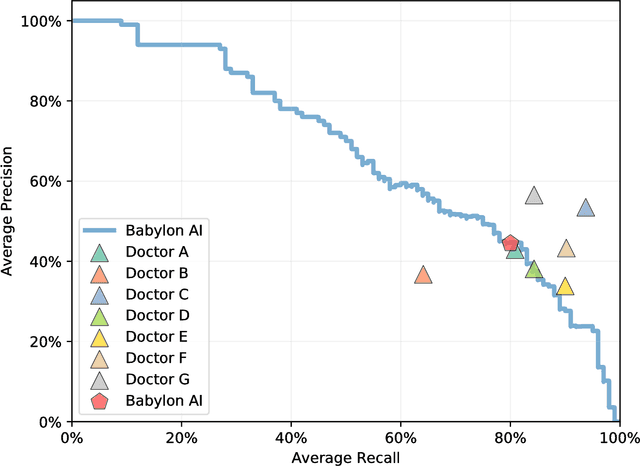
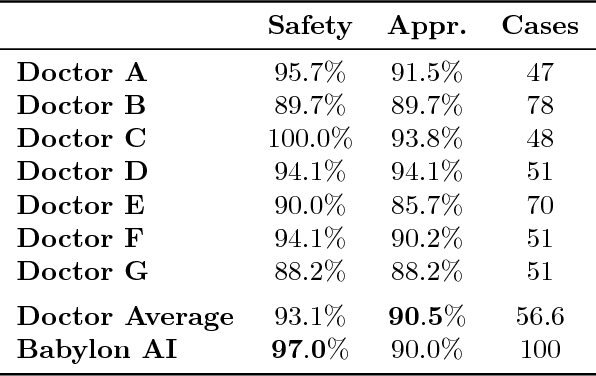
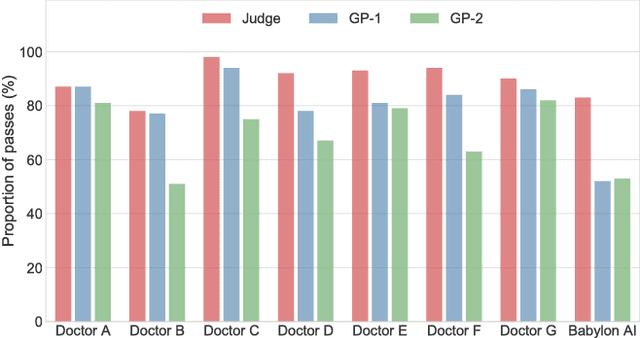
Abstract:Online symptom checkers have significant potential to improve patient care, however their reliability and accuracy remain variable. We hypothesised that an artificial intelligence (AI) powered triage and diagnostic system would compare favourably with human doctors with respect to triage and diagnostic accuracy. We performed a prospective validation study of the accuracy and safety of an AI powered triage and diagnostic system. Identical cases were evaluated by both an AI system and human doctors. Differential diagnoses and triage outcomes were evaluated by an independent judge, who was blinded from knowing the source (AI system or human doctor) of the outcomes. Independently of these cases, vignettes from publicly available resources were also assessed to provide a benchmark to previous studies and the diagnostic component of the MRCGP exam. Overall we found that the Babylon AI powered Triage and Diagnostic System was able to identify the condition modelled by a clinical vignette with accuracy comparable to human doctors (in terms of precision and recall). In addition, we found that the triage advice recommended by the AI System was, on average, safer than that of human doctors, when compared to the ranges of acceptable triage provided by independent expert judges, with only a minimal reduction in appropriateness.
A Universal Marginalizer for Amortized Inference in Generative Models
Nov 02, 2017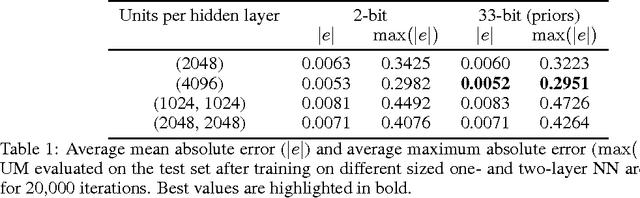
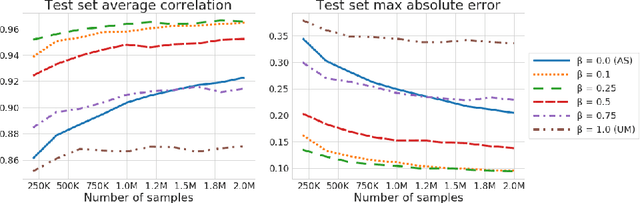
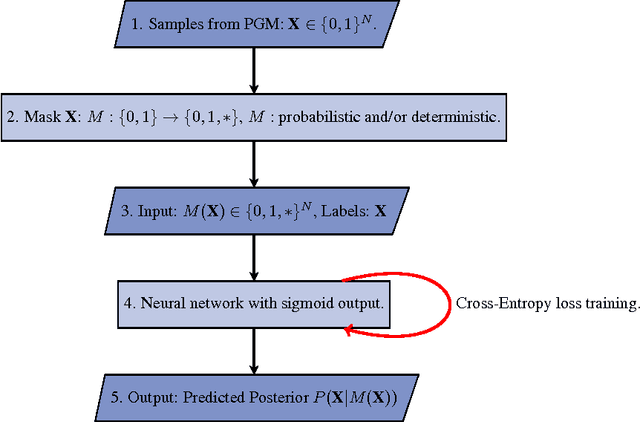
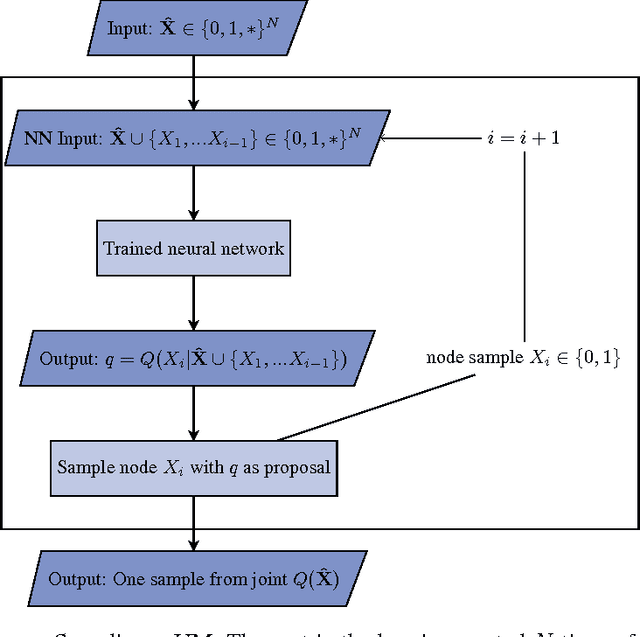
Abstract:We consider the problem of inference in a causal generative model where the set of available observations differs between data instances. We show how combining samples drawn from the graphical model with an appropriate masking function makes it possible to train a single neural network to approximate all the corresponding conditional marginal distributions and thus amortize the cost of inference. We further demonstrate that the efficiency of importance sampling may be improved by basing proposals on the output of the neural network. We also outline how the same network can be used to generate samples from an approximate joint posterior via a chain decomposition of the graph.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge