Saad Ullah Akram
Pedestrian Detection: Domain Generalization, CNNs, Transformers and Beyond
Jan 10, 2022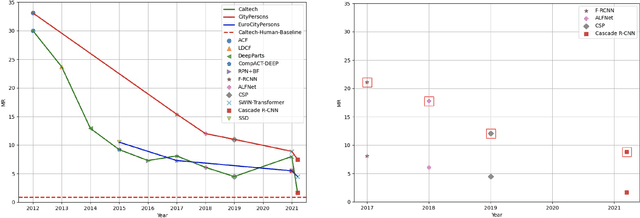

Abstract:Pedestrian detection is the cornerstone of many vision based applications, starting from object tracking to video surveillance and more recently, autonomous driving. With the rapid development of deep learning in object detection, pedestrian detection has achieved very good performance in traditional single-dataset training and evaluation setting. However, in this study on generalizable pedestrian detectors, we show that, current pedestrian detectors poorly handle even small domain shifts in cross-dataset evaluation. We attribute the limited generalization to two main factors, the method and the current sources of data. Regarding the method, we illustrate that biasness present in the design choices (e.g anchor settings) of current pedestrian detectors are the main contributing factor to the limited generalization. Most modern pedestrian detectors are tailored towards target dataset, where they do achieve high performance in traditional single training and testing pipeline, but suffer a degrade in performance when evaluated through cross-dataset evaluation. Consequently, a general object detector performs better in cross-dataset evaluation compared with state of the art pedestrian detectors, due to its generic design. As for the data, we show that the autonomous driving benchmarks are monotonous in nature, that is, they are not diverse in scenarios and dense in pedestrians. Therefore, benchmarks curated by crawling the web (which contain diverse and dense scenarios), are an efficient source of pre-training for providing a more robust representation. Accordingly, we propose a progressive fine-tuning strategy which improves generalization. Code and models cab accessed at https://github.com/hasanirtiza/Pedestron.
Bridging the gap between paired and unpaired medical image translation
Oct 15, 2021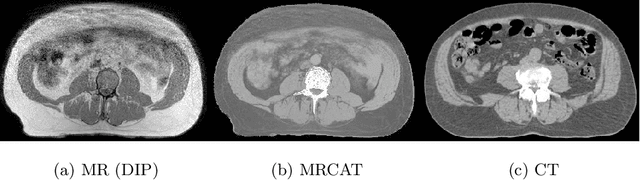

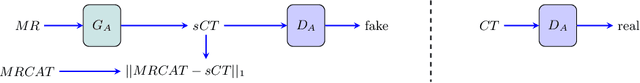
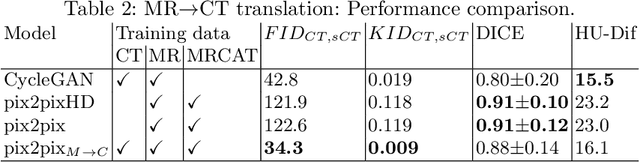
Abstract:Medical image translation has the potential to reduce the imaging workload, by removing the need to capture some sequences, and to reduce the annotation burden for developing machine learning methods. GANs have been used successfully to translate images from one domain to another, such as MR to CT. At present, paired data (registered MR and CT images) or extra supervision (e.g. segmentation masks) is needed to learn good translation models. Registering multiple modalities or annotating structures within each of them is a tedious and laborious task. Thus, there is a need to develop improved translation methods for unpaired data. Here, we introduce modified pix2pix models for tasks CT$\rightarrow$MR and MR$\rightarrow$CT, trained with unpaired CT and MR data, and MRCAT pairs generated from the MR scans. The proposed modifications utilize the paired MR and MRCAT images to ensure good alignment between input and translated images, and unpaired CT images ensure the MR$\rightarrow$CT model produces realistic-looking CT and CT$\rightarrow$MR model works well with real CT as input. The proposed pix2pix variants outperform baseline pix2pix, pix2pixHD and CycleGAN in terms of FID and KID, and generate more realistic looking CT and MR translations.
Pedestrian Detection: The Elephant In The Room
Mar 22, 2020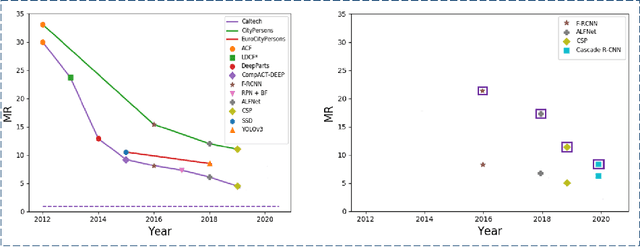
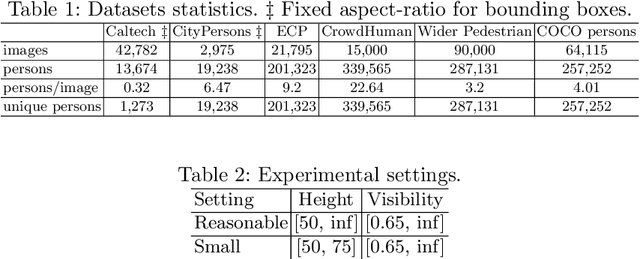

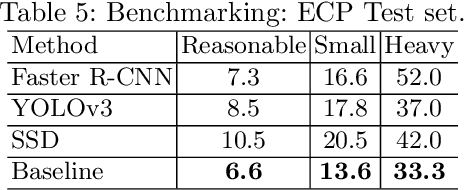
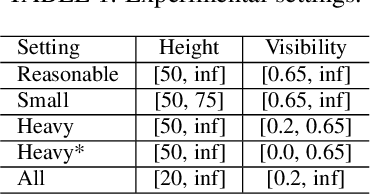
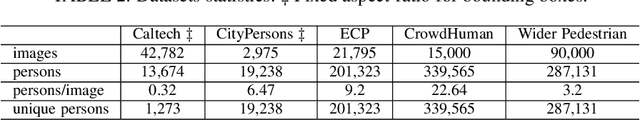
Abstract:Pedestrian detection is used in many vision based applications ranging from video surveillance to autonomous driving. Despite achieving high performance, it is still largely unknown how well existing detectors generalize to unseen data. To this end, we conduct a comprehensive study in this paper, using a general principle of direct cross-dataset evaluation. Through this study, we find that existing state-of-the-art pedestrian detectors generalize poorly from one dataset to another. We demonstrate that there are two reasons for this trend. Firstly, they over-fit on popular datasets in a traditional single-dataset training and test pipeline. Secondly, the training source is generally not dense in pedestrians and diverse in scenarios. Accordingly, through experiments we find that a general purpose object detector works better in direct cross-dataset evaluation compared with state-of-the-art pedestrian detectors and we illustrate that diverse and dense datasets, collected by crawling the web, serve to be an efficient source of pre-training for pedestrian detection. Furthermore, we find that a progressive training pipeline works good for autonomous driving oriented detector. We improve upon previous state-of-the-art on reasonable/heavy subsets of CityPersons dataset by 1.3%/1.7% and on Caltech by 1.8%/14.9% in terms of log average miss rate (MR^2) points without any fine-tuning on the test set. Detector trained through proposed pipeline achieves top rank at the leaderborads of CityPersons [42] and ECP [4]. Code and models will be available at https://github.com/hasanirtiza/Pedestron.
Mask-RCNN and U-net Ensembled for Nuclei Segmentation
Jan 29, 2019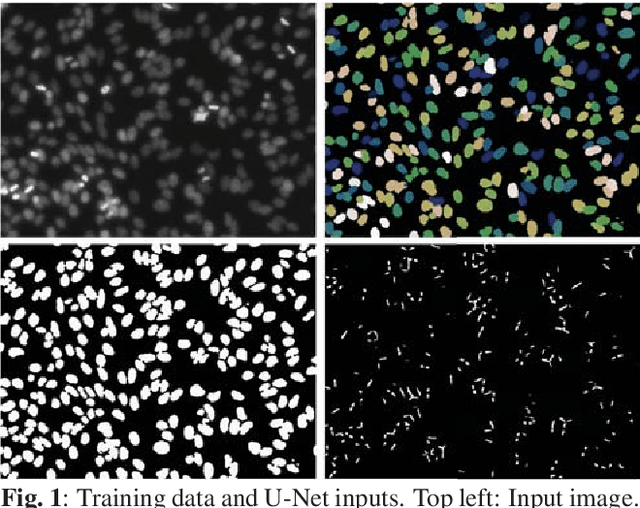
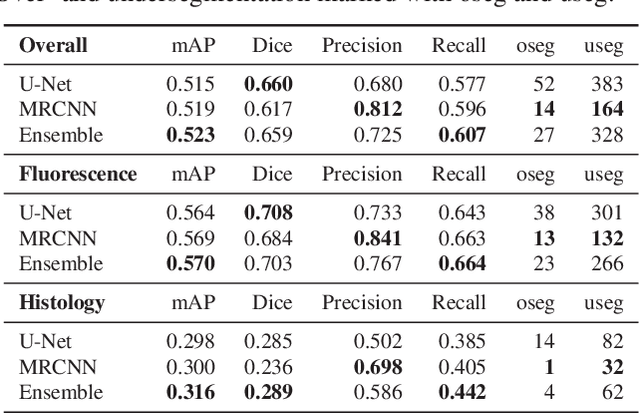
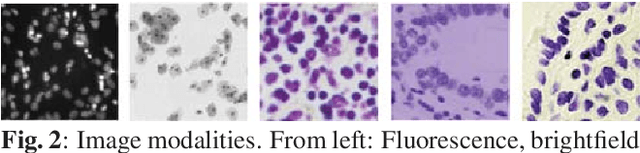

Abstract:Nuclei segmentation is both an important and in some ways ideal task for modern computer vision methods, e.g. convolutional neural networks. While recent developments in theory and open-source software have made these tools easier to implement, expert knowledge is still required to choose the right model architecture and training setup. We compare two popular segmentation frameworks, U-Net and Mask-RCNN in the nuclei segmentation task and find that they have different strengths and failures. To get the best of both worlds, we develop an ensemble model to combine their predictions that can outperform both models by a significant margin and should be considered when aiming for best nuclei segmentation performance.
Leveraging Unlabeled Whole-Slide-Images for Mitosis Detection
Jul 31, 2018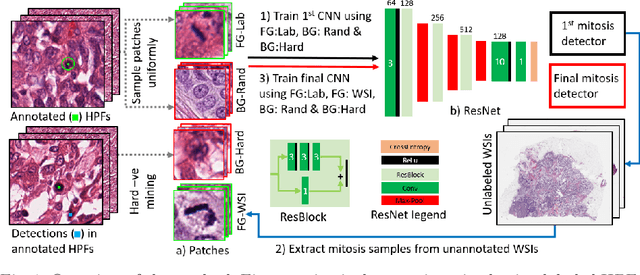
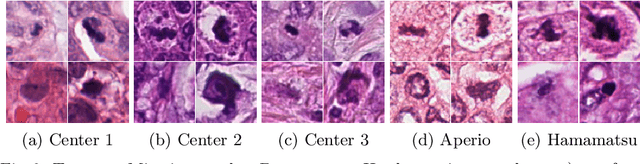
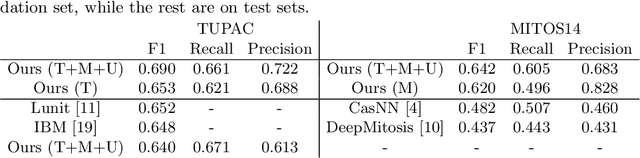
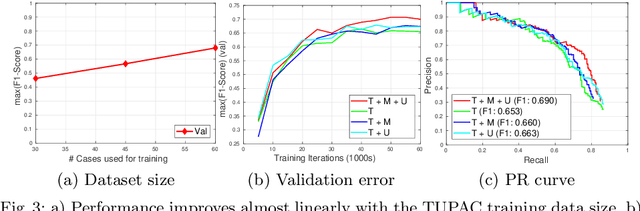
Abstract:Mitosis count is an important biomarker for prognosis of various cancers. At present, pathologists typically perform manual counting on a few selected regions of interest in breast whole-slide-images (WSIs) of patient biopsies. This task is very time-consuming, tedious and subjective. Automated mitosis detection methods have made great advances in recent years. However, these methods require exhaustive labeling of a large number of selected regions of interest. This task is very expensive because expert pathologists are needed for reliable and accurate annotations. In this paper, we present a semi-supervised mitosis detection method which is designed to leverage a large number of unlabeled breast cancer WSIs. As a result, our method capitalizes on the growing number of digitized histology images, without relying on exhaustive annotations, subsequently improving mitosis detection. Our method first learns a mitosis detector from labeled data, uses this detector to mine additional mitosis samples from unlabeled WSIs, and then trains the final model using this larger and diverse set of mitosis samples. The use of unlabeled data improves F1-score by $\sim$5\% compared to our best performing fully-supervised model on the TUPAC validation set. Our submission (single model) to TUPAC challenge ranks highly on the leaderboard with an F1-score of 0.64.
Cell Tracking via Proposal Generation and Selection
May 09, 2017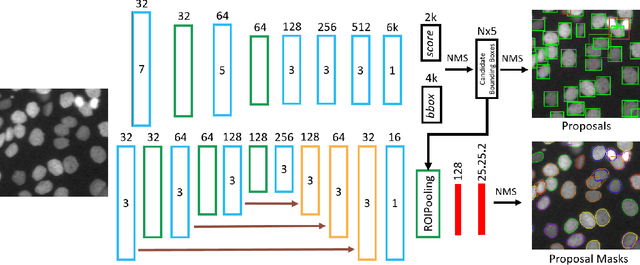
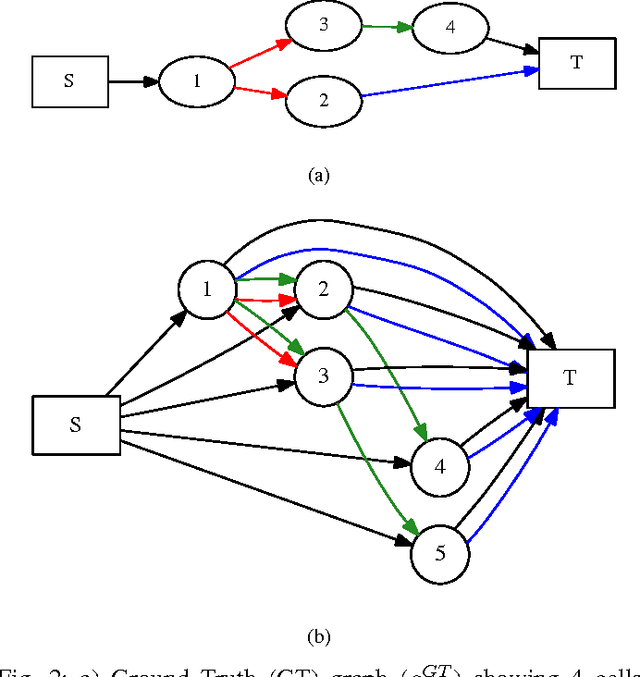
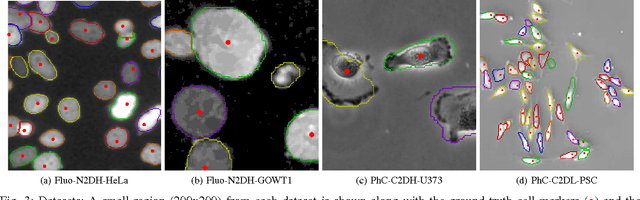
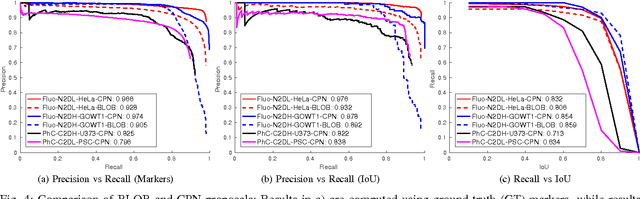
Abstract:Microscopy imaging plays a vital role in understanding many biological processes in development and disease. The recent advances in automation of microscopes and development of methods and markers for live cell imaging has led to rapid growth in the amount of image data being captured. To efficiently and reliably extract useful insights from these captured sequences, automated cell tracking is essential. This is a challenging problem due to large variation in the appearance and shapes of cells depending on many factors including imaging methodology, biological characteristics of cells, cell matrix composition, labeling methodology, etc. Often cell tracking methods require a sequence-specific segmentation method and manual tuning of many tracking parameters, which limits their applicability to sequences other than those they are designed for. In this paper, we propose 1) a deep learning based cell proposal method, which proposes candidates for cells along with their scores, and 2) a cell tracking method, which links proposals in adjacent frames in a graphical model using edges representing different cellular events and poses joint cell detection and tracking as the selection of a subset of cell and edge proposals. Our method is completely automated and given enough training data can be applied to a wide variety of microscopy sequences. We evaluate our method on multiple fluorescence and phase contrast microscopy sequences containing cells of various shapes and appearances from ISBI cell tracking challenge, and show that our method outperforms existing cell tracking methods. Code is available at: https://github.com/SaadUllahAkram/CellTracker
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge