Robert A. Barton
Predicting Cellular Responses with Variational Causal Inference and Refined Relational Information
Sep 30, 2022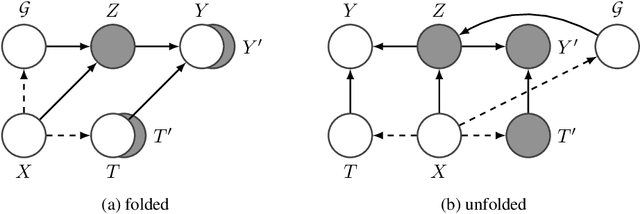
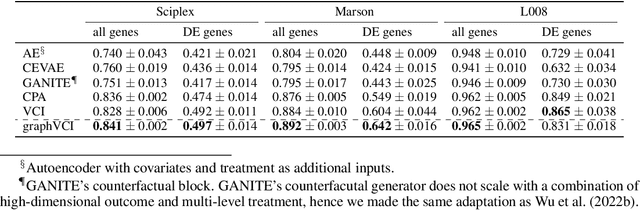
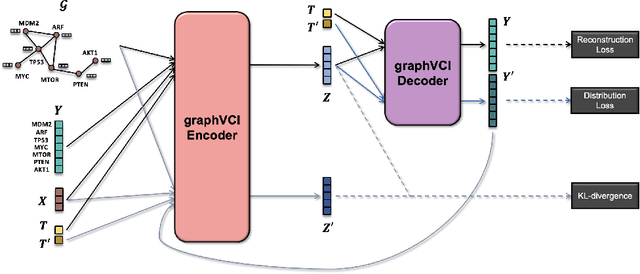
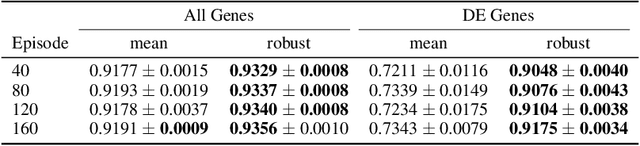
Abstract:Predicting the responses of a cell under perturbations may bring important benefits to drug discovery and personalized therapeutics. In this work, we propose a novel graph variational Bayesian causal inference framework to predict a cell's gene expressions under counterfactual perturbations (perturbations that this cell did not factually receive), leveraging information representing biological knowledge in the form of gene regulatory networks (GRNs) to aid individualized cellular response predictions. Aiming at a data-adaptive GRN, we also developed an adjacency matrix updating technique for graph convolutional networks and used it to refine GRNs during pre-training, which generated more insights on gene relations and enhanced model performance. Additionally, we propose a robust estimator within our framework for the asymptotically efficient estimation of marginal perturbation effect, which is yet to be carried out in previous works. With extensive experiments, we exhibited the advantage of our approach over state-of-the-art deep learning models for individual response prediction.
Neural Design for Genetic Perturbation Experiments
Jul 26, 2022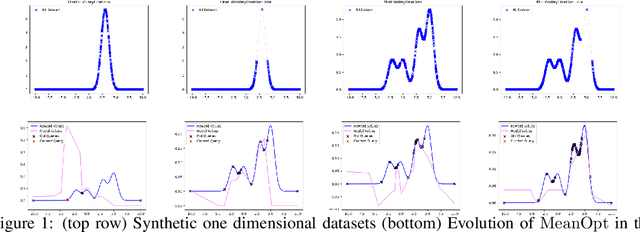
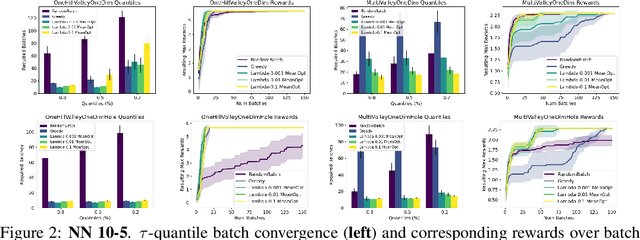
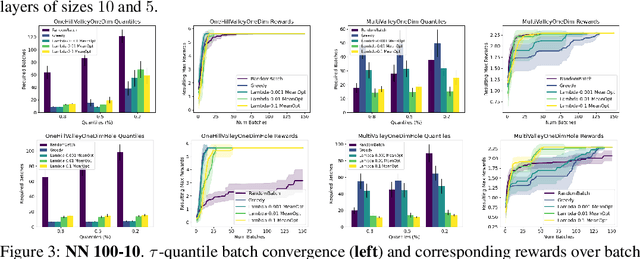
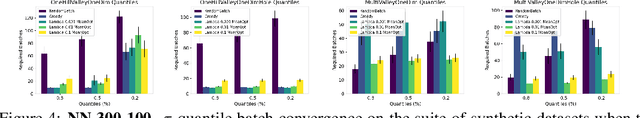
Abstract:The problem of how to genetically modify cells in order to maximize a certain cellular phenotype has taken center stage in drug development over the last few years (with, for example, genetically edited CAR-T, CAR-NK, and CAR-NKT cells entering cancer clinical trials). Exhausting the search space for all possible genetic edits (perturbations) or combinations thereof is infeasible due to cost and experimental limitations. This work provides a theoretically sound framework for iteratively exploring the space of perturbations in pooled batches in order to maximize a target phenotype under an experimental budget. Inspired by this application domain, we study the problem of batch query bandit optimization and introduce the Optimistic Arm Elimination ($\mathrm{OAE}$) principle designed to find an almost optimal arm under different functional relationships between the queries (arms) and the outputs (rewards). We analyze the convergence properties of $\mathrm{OAE}$ by relating it to the Eluder dimension of the algorithm's function class and validate that $\mathrm{OAE}$ outperforms other strategies in finding optimal actions in experiments on simulated problems, public datasets well-studied in bandit contexts, and in genetic perturbation datasets when the regression model is a deep neural network. OAE also outperforms the benchmark algorithms in 3 of 4 datasets in the GeneDisco experimental planning challenge.
SystemMatch: optimizing preclinical drug models to human clinical outcomes via generative latent-space matching
May 14, 2022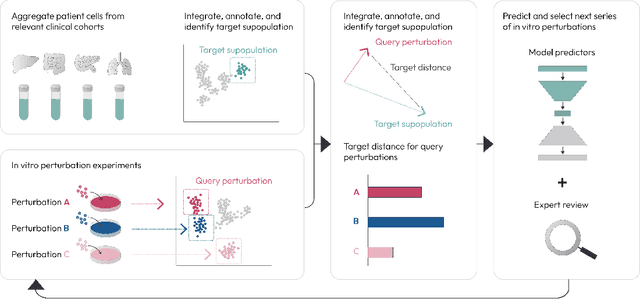
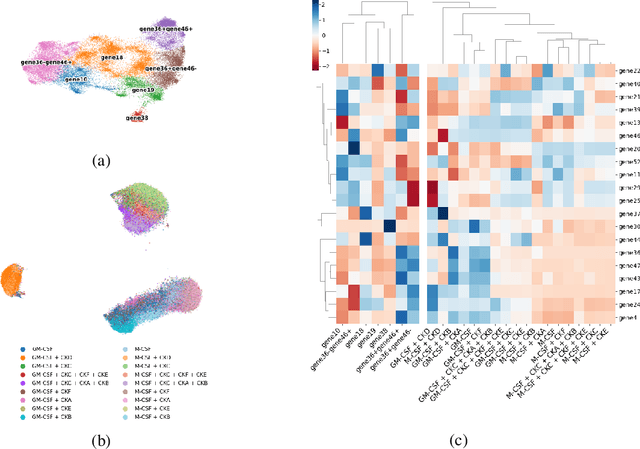
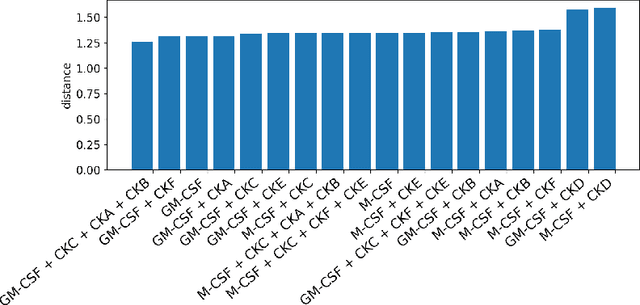
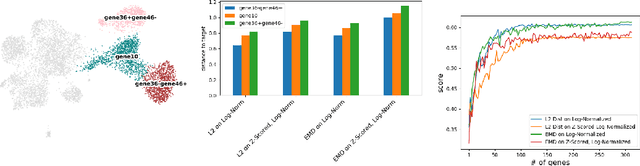
Abstract:Translating the relevance of preclinical models ($\textit{in vitro}$, animal models, or organoids) to their relevance in humans presents an important challenge during drug development. The rising abundance of single-cell genomic data from human tumors and tissue offers a new opportunity to optimize model systems by their similarity to targeted human cell types in disease. In this work, we introduce SystemMatch to assess the fit of preclinical model systems to an $\textit{in sapiens}$ target population and to recommend experimental changes to further optimize these systems. We demonstrate this through an application to developing $\textit{in vitro}$ systems to model human tumor-derived suppressive macrophages. We show with held-out $\textit{in vivo}$ controls that our pipeline successfully ranks macrophage subpopulations by their biological similarity to the target population, and apply this analysis to rank a series of 18 $\textit{in vitro}$ macrophage systems perturbed with a variety of cytokine stimulations. We extend this analysis to predict the behavior of 66 $\textit{in silico}$ model systems generated using a perturbational autoencoder and apply a $k$-medoids approach to recommend a subset of these model systems for further experimental development in order to fully explore the space of possible perturbations. Through this use case, we demonstrate a novel approach to model system development to generate a system more similar to human biology.
Graph Neural Networks for Inconsistent Cluster Detection in Incremental Entity Resolution
May 12, 2021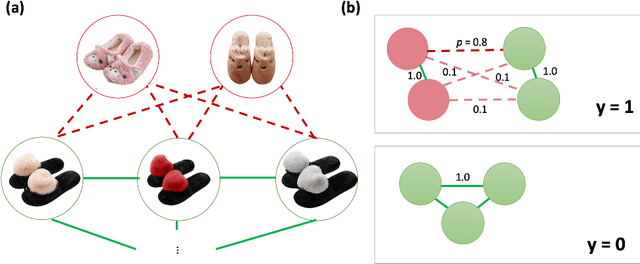
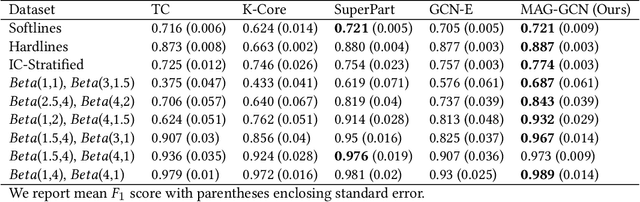
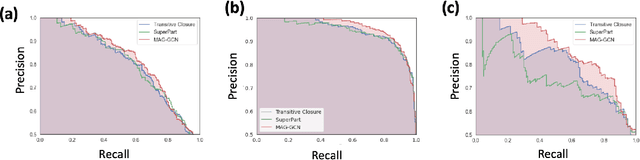
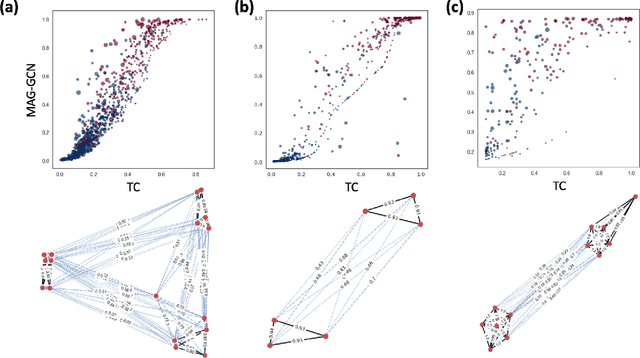
Abstract:Online stores often utilize product relationships such as bundles and substitutes to improve their catalog quality and guide customers through myriad choices. Entity resolution using pairwise product matching models offers a means of inferring relationships between products. In mature data repositories, the relationships may be mostly correct but require incremental improvements owing to errors in the original data or in the entity resolution system. It is critical to devise incremental entity resolution (IER) approaches for improving the health of relationships. However, most existing research on IER focuses on the addition of new products or information into existing relationships. Relatively little research has been done for detecting low quality within current relationships. This paper proposes a novel method for identifying inconsistent clusters (IC), existing groups of related products that do not belong together. We propose to treat the identification of inconsistent clusters as a supervised learning task which predicts whether a graph of products with similarities as weighted edges should be partitioned into multiple clusters. In this case, the problem becomes a classification task on weighted graphs and represents an interesting application area for modern tools such as Graph Neural Networks (GNNs). We demonstrate that existing Message Passing neural networks perform well at this task, exceeding traditional graph processing techniques. We also develop a novel message aggregation scheme for Message Passing Neural Networks that further improves the performance of GNNs on this task. We apply the model to synthetic datasets, a public benchmark dataset, and an internal application. Our results demonstrate the value of graph classification in IER and the ability of graph neural networks to develop useful representations for graph partitioning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge