Luis F. Voloch
Predicting Cellular Responses with Variational Causal Inference and Refined Relational Information
Sep 30, 2022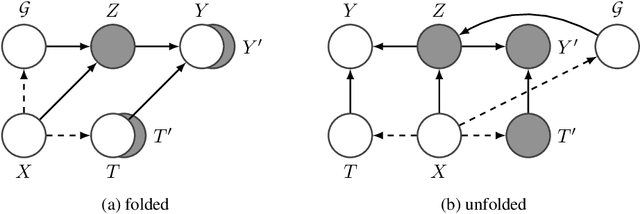
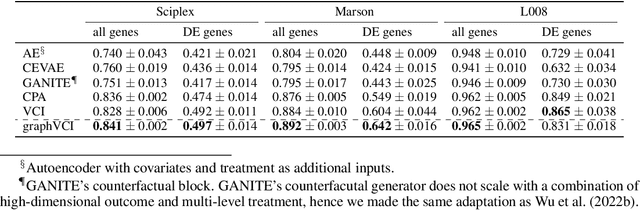
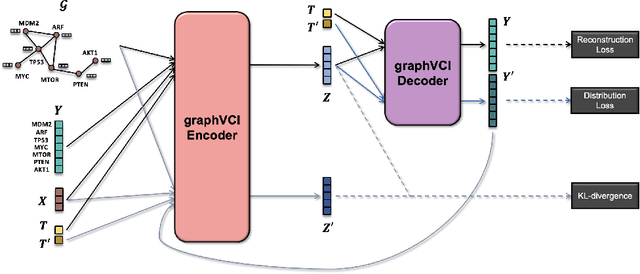
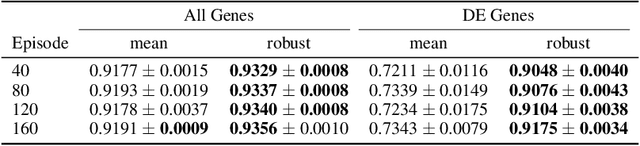
Abstract:Predicting the responses of a cell under perturbations may bring important benefits to drug discovery and personalized therapeutics. In this work, we propose a novel graph variational Bayesian causal inference framework to predict a cell's gene expressions under counterfactual perturbations (perturbations that this cell did not factually receive), leveraging information representing biological knowledge in the form of gene regulatory networks (GRNs) to aid individualized cellular response predictions. Aiming at a data-adaptive GRN, we also developed an adjacency matrix updating technique for graph convolutional networks and used it to refine GRNs during pre-training, which generated more insights on gene relations and enhanced model performance. Additionally, we propose a robust estimator within our framework for the asymptotically efficient estimation of marginal perturbation effect, which is yet to be carried out in previous works. With extensive experiments, we exhibited the advantage of our approach over state-of-the-art deep learning models for individual response prediction.
SystemMatch: optimizing preclinical drug models to human clinical outcomes via generative latent-space matching
May 14, 2022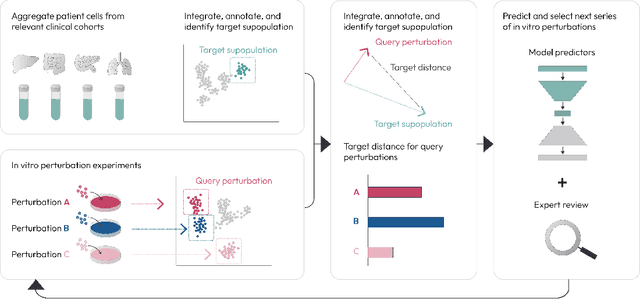
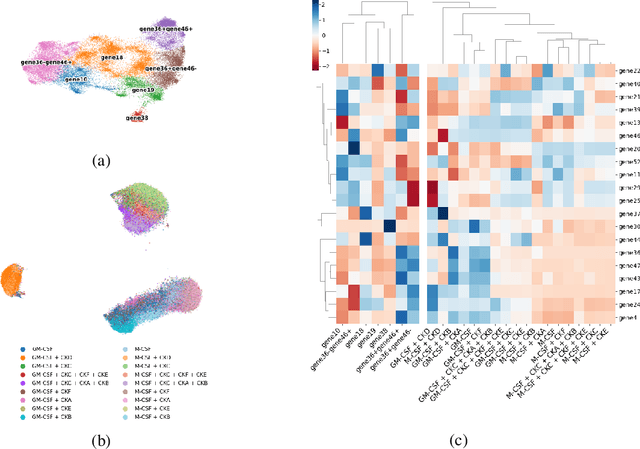
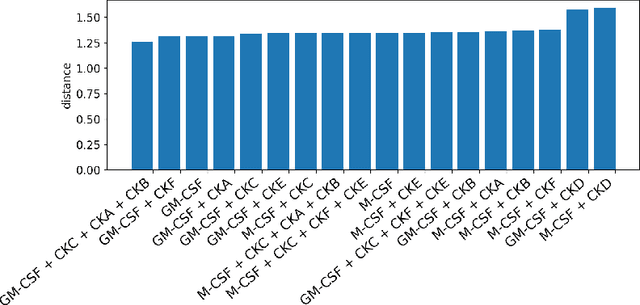
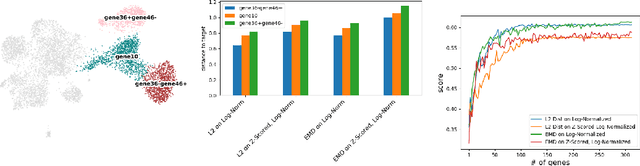
Abstract:Translating the relevance of preclinical models ($\textit{in vitro}$, animal models, or organoids) to their relevance in humans presents an important challenge during drug development. The rising abundance of single-cell genomic data from human tumors and tissue offers a new opportunity to optimize model systems by their similarity to targeted human cell types in disease. In this work, we introduce SystemMatch to assess the fit of preclinical model systems to an $\textit{in sapiens}$ target population and to recommend experimental changes to further optimize these systems. We demonstrate this through an application to developing $\textit{in vitro}$ systems to model human tumor-derived suppressive macrophages. We show with held-out $\textit{in vivo}$ controls that our pipeline successfully ranks macrophage subpopulations by their biological similarity to the target population, and apply this analysis to rank a series of 18 $\textit{in vitro}$ macrophage systems perturbed with a variety of cytokine stimulations. We extend this analysis to predict the behavior of 66 $\textit{in silico}$ model systems generated using a perturbational autoencoder and apply a $k$-medoids approach to recommend a subset of these model systems for further experimental development in order to fully explore the space of possible perturbations. Through this use case, we demonstrate a novel approach to model system development to generate a system more similar to human biology.
Regret Guarantees for Item-Item Collaborative Filtering
Jan 08, 2016
Abstract:There is much empirical evidence that item-item collaborative filtering works well in practice. Motivated to understand this, we provide a framework to design and analyze various recommendation algorithms. The setup amounts to online binary matrix completion, where at each time a random user requests a recommendation and the algorithm chooses an entry to reveal in the user's row. The goal is to minimize regret, or equivalently to maximize the number of +1 entries revealed at any time. We analyze an item-item collaborative filtering algorithm that can achieve fundamentally better performance compared to user-user collaborative filtering. The algorithm achieves good "cold-start" performance (appropriately defined) by quickly making good recommendations to new users about whom there is little information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge