Pietro Mercati
LogHD: Robust Compression of Hyperdimensional Classifiers via Logarithmic Class-Axis Reduction
Nov 06, 2025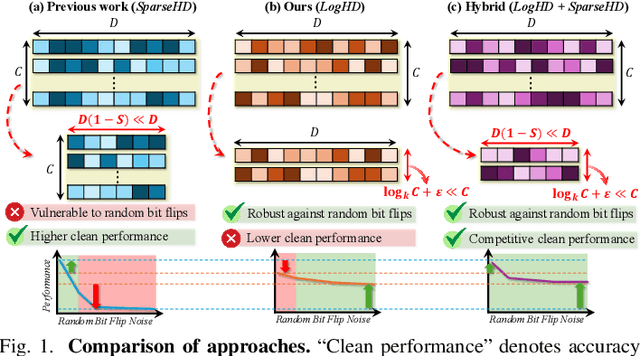
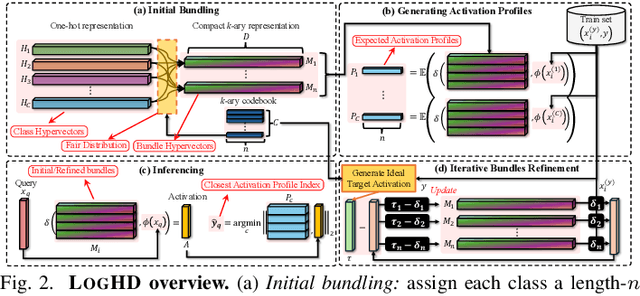
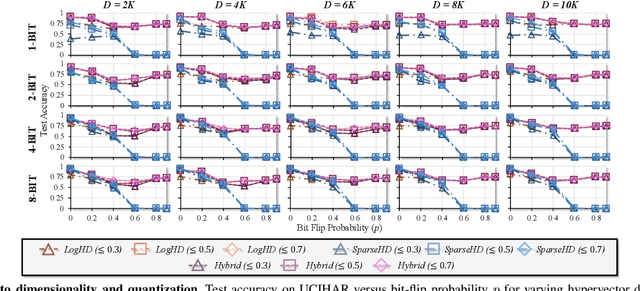
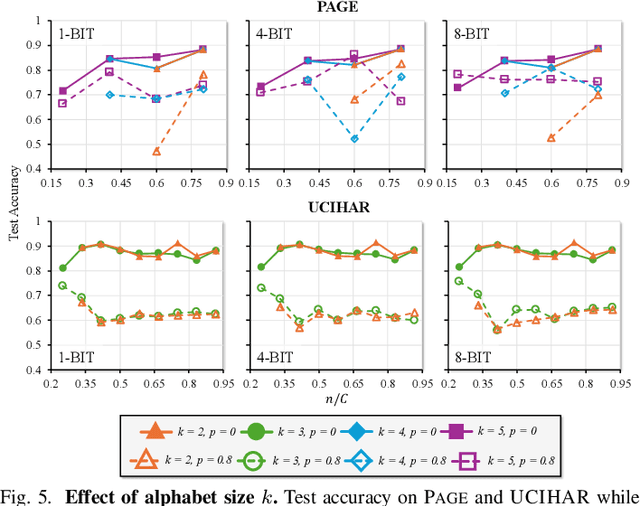
Abstract:Hyperdimensional computing (HDC) suits memory, energy, and reliability-constrained systems, yet the standard "one prototype per class" design requires $O(CD)$ memory (with $C$ classes and dimensionality $D$). Prior compaction reduces $D$ (feature axis), improving storage/compute but weakening robustness. We introduce LogHD, a logarithmic class-axis reduction that replaces the $C$ per-class prototypes with $n\!\approx\!\lceil\log_k C\rceil$ bundle hypervectors (alphabet size $k$) and decodes in an $n$-dimensional activation space, cutting memory to $O(D\log_k C)$ while preserving $D$. LogHD uses a capacity-aware codebook and profile-based decoding, and composes with feature-axis sparsification. Across datasets and injected bit flips, LogHD attains competitive accuracy with smaller models and higher resilience at matched memory. Under equal memory, it sustains target accuracy at roughly $2.5$-$3.0\times$ higher bit-flip rates than feature-axis compression; an ASIC instantiation delivers $498\times$ energy efficiency and $62.6\times$ speedup over an AMD Ryzen 9 9950X and $24.3\times$/$6.58\times$ over an NVIDIA RTX 4090, and is $4.06\times$ more energy-efficient and $2.19\times$ faster than a feature-axis HDC ASIC baseline.
HEAL: Brain-inspired Hyperdimensional Efficient Active Learning
Feb 17, 2024



Abstract:Drawing inspiration from the outstanding learning capability of our human brains, Hyperdimensional Computing (HDC) emerges as a novel computing paradigm, and it leverages high-dimensional vector presentation and operations for brain-like lightweight Machine Learning (ML). Practical deployments of HDC have significantly enhanced the learning efficiency compared to current deep ML methods on a broad spectrum of applications. However, boosting the data efficiency of HDC classifiers in supervised learning remains an open question. In this paper, we introduce Hyperdimensional Efficient Active Learning (HEAL), a novel Active Learning (AL) framework tailored for HDC classification. HEAL proactively annotates unlabeled data points via uncertainty and diversity-guided acquisition, leading to a more efficient dataset annotation and lowering labor costs. Unlike conventional AL methods that only support classifiers built upon deep neural networks (DNN), HEAL operates without the need for gradient or probabilistic computations. This allows it to be effortlessly integrated with any existing HDC classifier architecture. The key design of HEAL is a novel approach for uncertainty estimation in HDC classifiers through a lightweight HDC ensemble with prior hypervectors. Additionally, by exploiting hypervectors as prototypes (i.e., compact representations), we develop an extra metric for HEAL to select diverse samples within each batch for annotation. Our evaluation shows that HEAL surpasses a diverse set of baselines in AL quality and achieves notably faster acquisition than many BNN-powered or diversity-guided AL methods, recording 11 times to 40,000 times speedup in acquisition runtime per batch.
On the Benefits of Leveraging Structural Information in Planning Over the Learned Model
Mar 15, 2023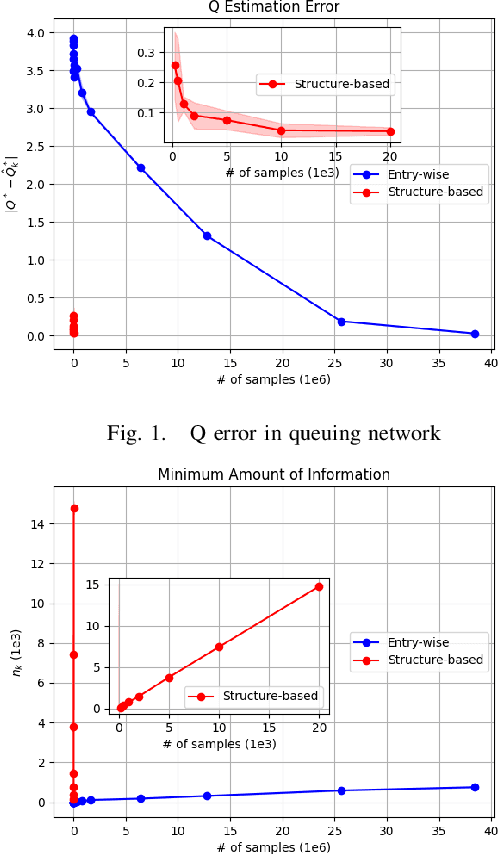
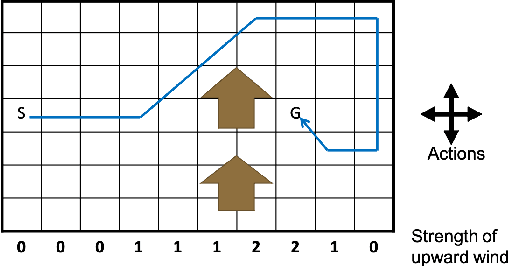
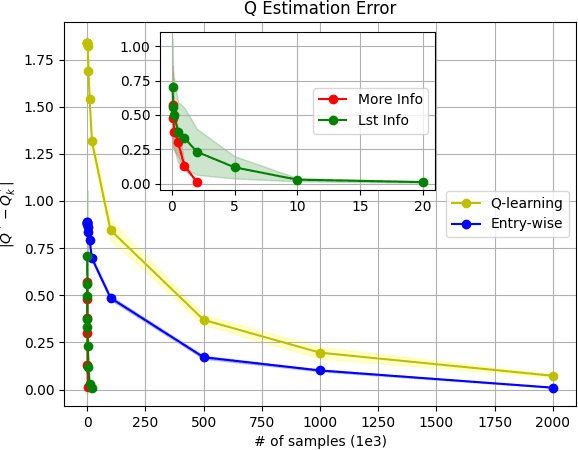
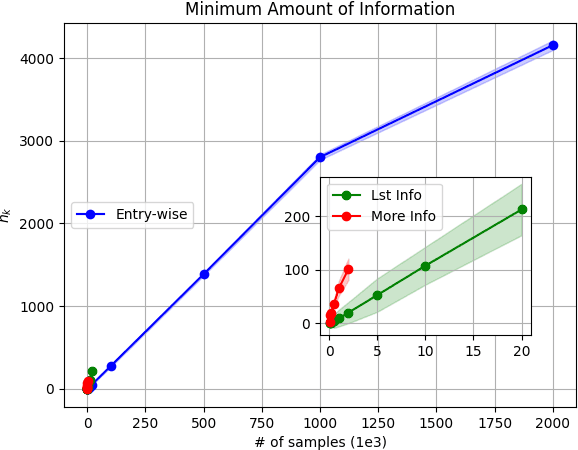
Abstract:Model-based Reinforcement Learning (RL) integrates learning and planning and has received increasing attention in recent years. However, learning the model can incur a significant cost (in terms of sample complexity), due to the need to obtain a sufficient number of samples for each state-action pair. In this paper, we investigate the benefits of leveraging structural information about the system in terms of reducing sample complexity. Specifically, we consider the setting where the transition probability matrix is a known function of a number of structural parameters, whose values are initially unknown. We then consider the problem of estimating those parameters based on the interactions with the environment. We characterize the difference between the Q estimates and the optimal Q value as a function of the number of samples. Our analysis shows that there can be a significant saving in sample complexity by leveraging structural information about the model. We illustrate the findings by considering several problems including controlling a queuing system with heterogeneous servers, and seeking an optimal path in a stochastic windy gridworld.
Automatic Tuning of Tensorflow's CPU Backend using Gradient-Free Optimization Algorithms
Sep 13, 2021



Abstract:Modern deep learning (DL) applications are built using DL libraries and frameworks such as TensorFlow and PyTorch. These frameworks have complex parameters and tuning them to obtain good training and inference performance is challenging for typical users, such as DL developers and data scientists. Manual tuning requires deep knowledge of the user-controllable parameters of DL frameworks as well as the underlying hardware. It is a slow and tedious process, and it typically delivers sub-optimal solutions. In this paper, we treat the problem of tuning parameters of DL frameworks to improve training and inference performance as a black-box optimization problem. We then investigate applicability and effectiveness of Bayesian optimization (BO), genetic algorithm (GA), and Nelder-Mead simplex (NMS) to tune the parameters of TensorFlow's CPU backend. While prior work has already investigated the use of Nelder-Mead simplex for a similar problem, it does not provide insights into the applicability of other more popular algorithms. Towards that end, we provide a systematic comparative analysis of all three algorithms in tuning TensorFlow's CPU backend on a variety of DL models. Our findings reveal that Bayesian optimization performs the best on the majority of models. There are, however, cases where it does not deliver the best results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge