Pascal Sturmfels
FoggySight: A Scheme for Facial Lookup Privacy
Dec 15, 2020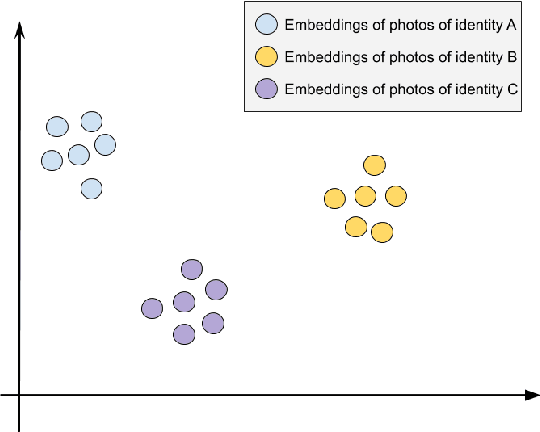
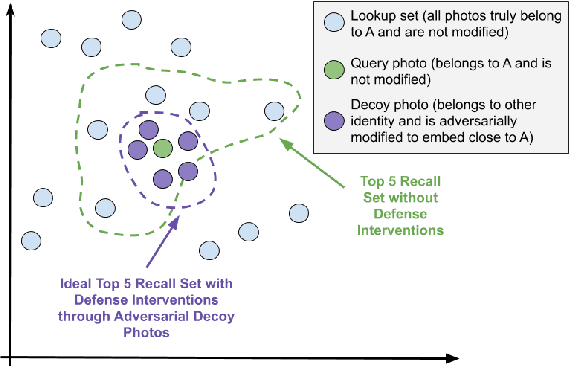
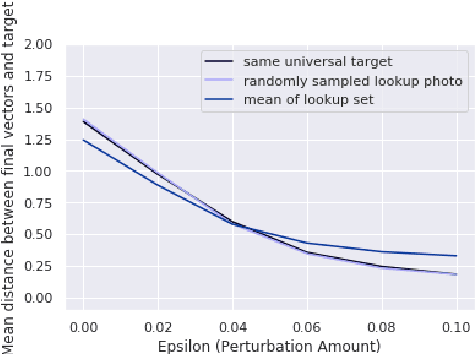

Abstract:Advances in deep learning algorithms have enabled better-than-human performance on face recognition tasks. In parallel, private companies have been scraping social media and other public websites that tie photos to identities and have built up large databases of labeled face images. Searches in these databases are now being offered as a service to law enforcement and others and carry a multitude of privacy risks for social media users. In this work, we tackle the problem of providing privacy from such face recognition systems. We propose and evaluate FoggySight, a solution that applies lessons learned from the adversarial examples literature to modify facial photos in a privacy-preserving manner before they are uploaded to social media. FoggySight's core feature is a community protection strategy where users acting as protectors of privacy for others upload decoy photos generated by adversarial machine learning algorithms. We explore different settings for this scheme and find that it does enable protection of facial privacy -- including against a facial recognition service with unknown internals.
Profile Prediction: An Alignment-Based Pre-Training Task for Protein Sequence Models
Dec 01, 2020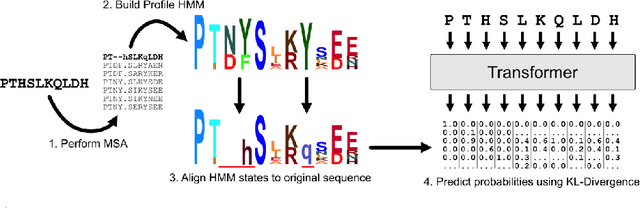
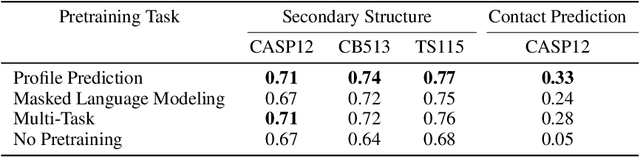
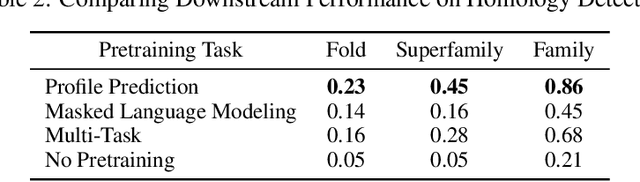
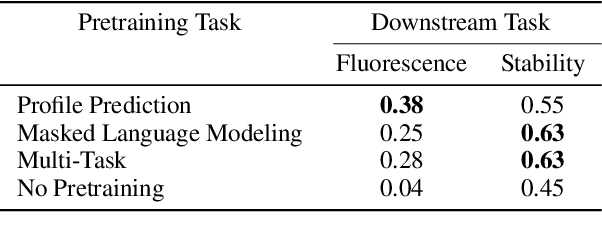
Abstract:For protein sequence datasets, unlabeled data has greatly outpaced labeled data due to the high cost of wet-lab characterization. Recent deep-learning approaches to protein prediction have shown that pre-training on unlabeled data can yield useful representations for downstream tasks. However, the optimal pre-training strategy remains an open question. Instead of strictly borrowing from natural language processing (NLP) in the form of masked or autoregressive language modeling, we introduce a new pre-training task: directly predicting protein profiles derived from multiple sequence alignments. Using a set of five, standardized downstream tasks for protein models, we demonstrate that our pre-training task along with a multi-task objective outperforms masked language modeling alone on all five tasks. Our results suggest that protein sequence models may benefit from leveraging biologically-inspired inductive biases that go beyond existing language modeling techniques in NLP.
Explaining Explanations: Axiomatic Feature Interactions for Deep Networks
Feb 12, 2020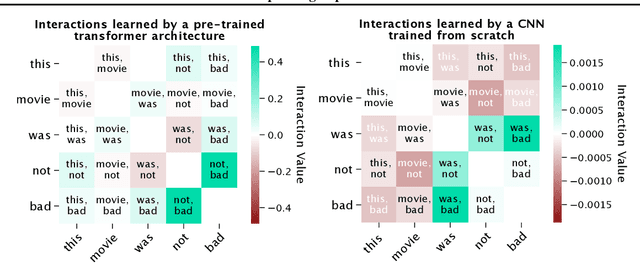
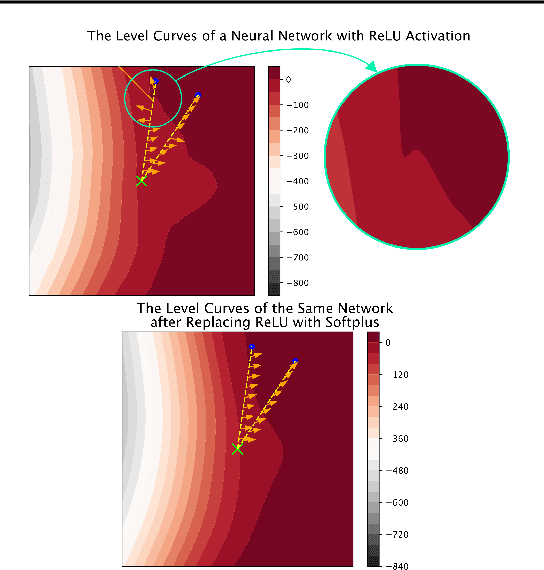

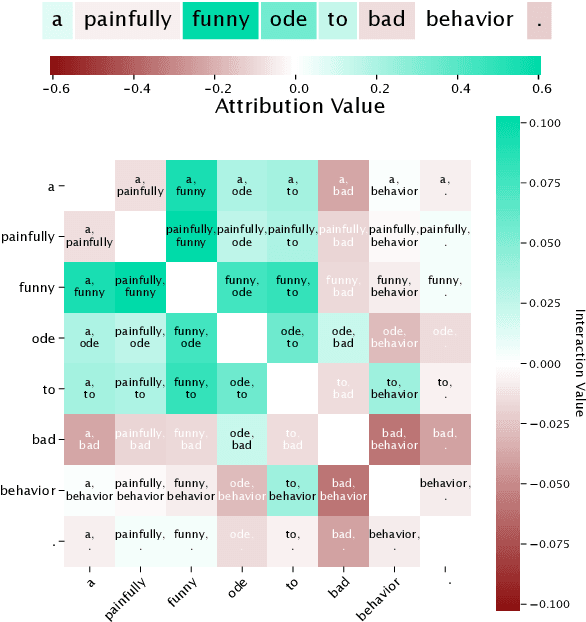
Abstract:Recent work has shown great promise in explaining neural network behavior. In particular, feature attribution methods explain which features were most important to a model's prediction on a given input. However, for many tasks, simply knowing which features were important to a model's prediction may not provide enough insight to understand model behavior. The interactions between features within the model may better help us understand not only the model, but also why certain features are more important than others. In this work we present Integrated Hessians, an extension of Integrated Gradients that explains pairwise feature interactions in neural networks. Integrated Hessians overcomes several theoretical limitations of previous methods to explain interactions, and unlike such previous methods is not limited to a specific architecture or class of neural network. We apply Integrated Hessians on a variety of neural networks trained on language data, biological data, astronomy data, and medical data and gain new insight into model behavior in each domain. Code available at https://github.com/suinleelab/path_explain
Learning Explainable Models Using Attribution Priors
Jun 25, 2019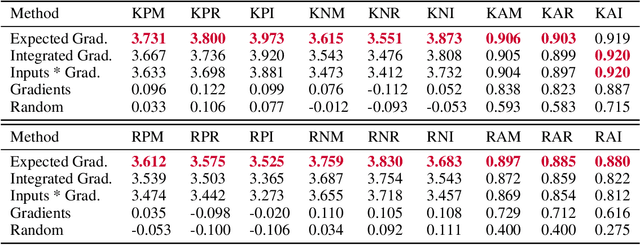

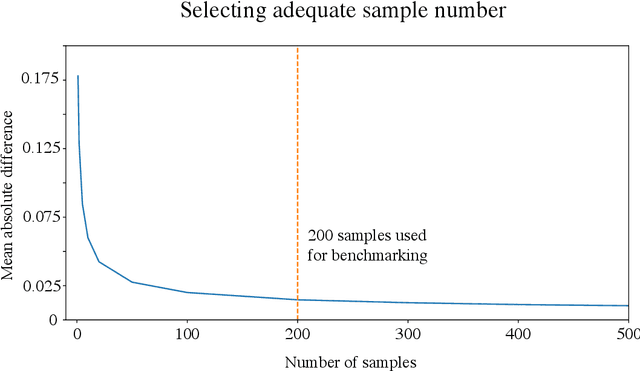
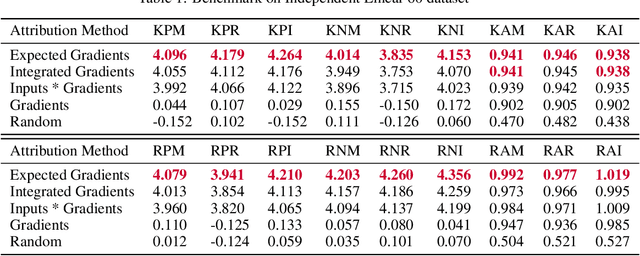
Abstract:Two important topics in deep learning both involve incorporating humans into the modeling process: Model priors transfer information from humans to a model by constraining the model's parameters; Model attributions transfer information from a model to humans by explaining the model's behavior. We propose connecting these topics with attribution priors (https://github.com/suinleelab/attributionpriors), which allow humans to use the common language of attributions to enforce prior expectations about a model's behavior during training. We develop a differentiable axiomatic feature attribution method called expected gradients and show how to directly regularize these attributions during training. We demonstrate the broad applicability of attribution priors ($\Omega$) by presenting three distinct examples that regularize models to behave more intuitively in three different domains: 1) on image data, $\Omega_{\textrm{pixel}}$ encourages models to have piecewise smooth attribution maps; 2) on gene expression data, $\Omega_{\textrm{graph}}$ encourages models to treat functionally related genes similarly; 3) on a health care dataset, $\Omega_{\textrm{sparse}}$ encourages models to rely on fewer features. In all three domains, attribution priors produce models with more intuitive behavior and better generalization performance by encoding constraints that would otherwise be very difficult to encode using standard model priors.
A Domain Guided CNN Architecture for Predicting Age from Structural Brain Images
Aug 11, 2018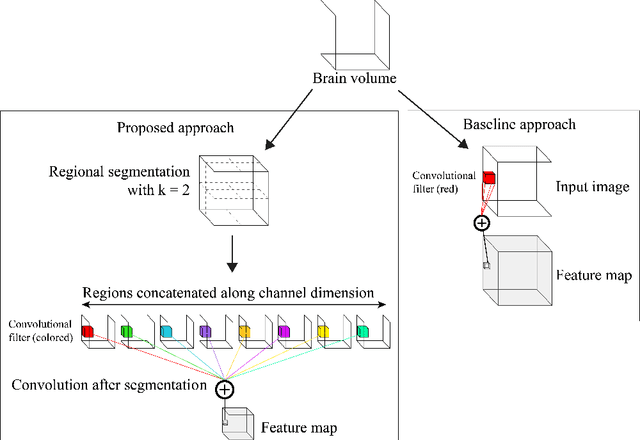
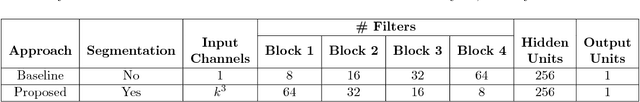
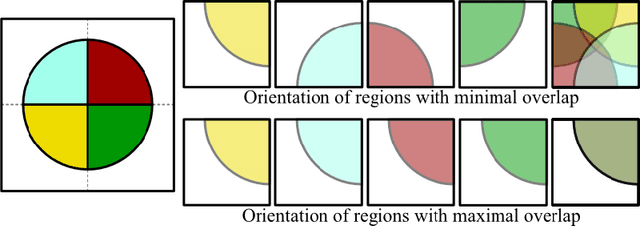
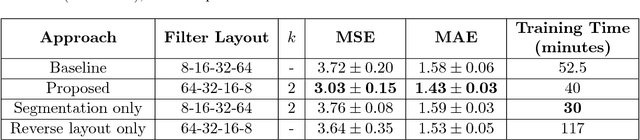
Abstract:Given the wide success of convolutional neural networks (CNNs) applied to natural images, researchers have begun to apply them to neuroimaging data. To date, however, exploration of novel CNN architectures tailored to neuroimaging data has been limited. Several recent works fail to leverage the 3D structure of the brain, instead treating the brain as a set of independent 2D slices. Approaches that do utilize 3D convolutions rely on architectures developed for object recognition tasks in natural 2D images. Such architectures make assumptions about the input that may not hold for neuroimaging. For example, existing architectures assume that patterns in the brain exhibit translation invariance. However, a pattern in the brain may have different meaning depending on where in the brain it is located. There is a need to explore novel architectures that are tailored to brain images. We present two simple modifications to existing CNN architectures based on brain image structure. Applied to the task of brain age prediction, our network achieves a mean absolute error (MAE) of 1.4 years and trains 30% faster than a CNN baseline that achieves a MAE of 1.6 years. Our results suggest that lessons learned from developing models on natural images may not directly transfer to neuroimaging tasks. Instead, there remains a large space of unexplored questions regarding model development in this area, whose answers may differ from conventional wisdom.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge