Gabriel Erion
An Adversarial Approach for the Robust Classification of Pneumonia from Chest Radiographs
Jan 13, 2020
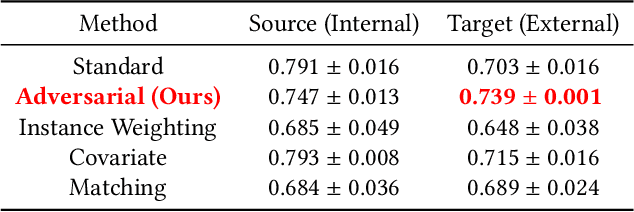
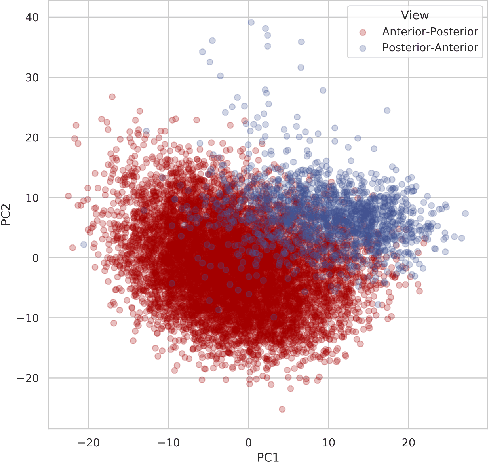
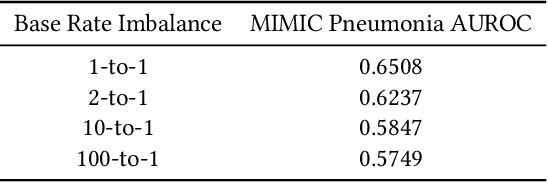
Abstract:While deep learning has shown promise in the domain of disease classification from medical images, models based on state-of-the-art convolutional neural network architectures often exhibit performance loss due to dataset shift. Models trained using data from one hospital system achieve high predictive performance when tested on data from the same hospital, but perform significantly worse when they are tested in different hospital systems. Furthermore, even within a given hospital system, deep learning models have been shown to depend on hospital- and patient-level confounders rather than meaningful pathology to make classifications. In order for these models to be safely deployed, we would like to ensure that they do not use confounding variables to make their classification, and that they will work well even when tested on images from hospitals that were not included in the training data. We attempt to address this problem in the context of pneumonia classification from chest radiographs. We propose an approach based on adversarial optimization, which allows us to learn more robust models that do not depend on confounders. Specifically, we demonstrate improved out-of-hospital generalization performance of a pneumonia classifier by training a model that is invariant to the view position of chest radiographs (anterior-posterior vs. posterior-anterior). Our approach leads to better predictive performance on external hospital data than both a standard baseline and previously proposed methods to handle confounding, and also suggests a method for identifying models that may rely on confounders. Code available at https://github.com/suinleelab/cxr_adv.
Learning Explainable Models Using Attribution Priors
Jun 25, 2019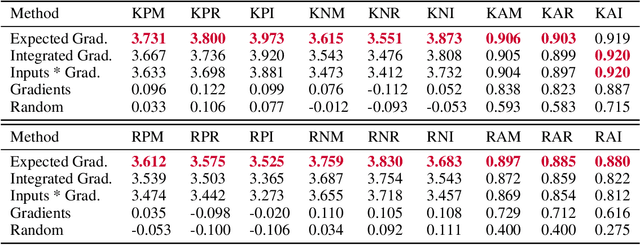

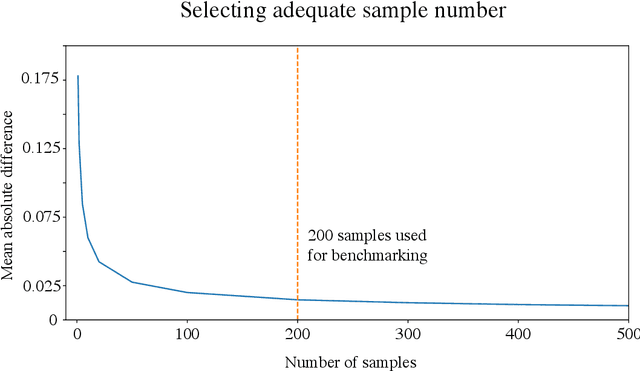
Abstract:Two important topics in deep learning both involve incorporating humans into the modeling process: Model priors transfer information from humans to a model by constraining the model's parameters; Model attributions transfer information from a model to humans by explaining the model's behavior. We propose connecting these topics with attribution priors (https://github.com/suinleelab/attributionpriors), which allow humans to use the common language of attributions to enforce prior expectations about a model's behavior during training. We develop a differentiable axiomatic feature attribution method called expected gradients and show how to directly regularize these attributions during training. We demonstrate the broad applicability of attribution priors ($\Omega$) by presenting three distinct examples that regularize models to behave more intuitively in three different domains: 1) on image data, $\Omega_{\textrm{pixel}}$ encourages models to have piecewise smooth attribution maps; 2) on gene expression data, $\Omega_{\textrm{graph}}$ encourages models to treat functionally related genes similarly; 3) on a health care dataset, $\Omega_{\textrm{sparse}}$ encourages models to rely on fewer features. In all three domains, attribution priors produce models with more intuitive behavior and better generalization performance by encoding constraints that would otherwise be very difficult to encode using standard model priors.
Explainable AI for Trees: From Local Explanations to Global Understanding
May 11, 2019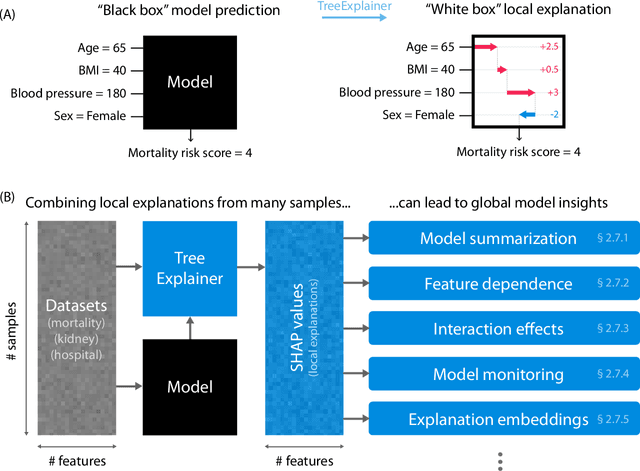
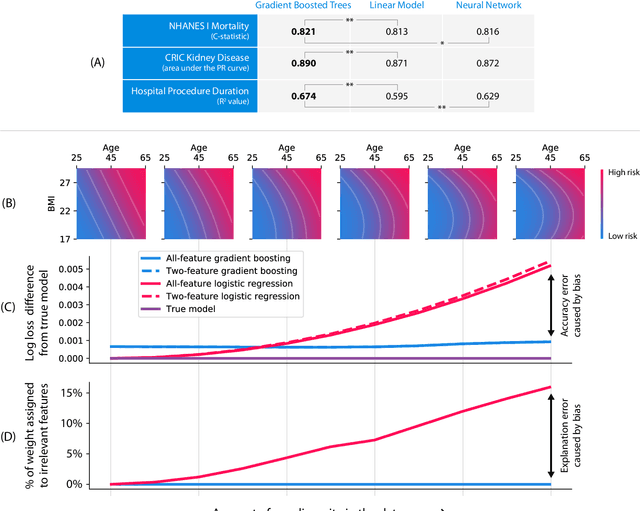
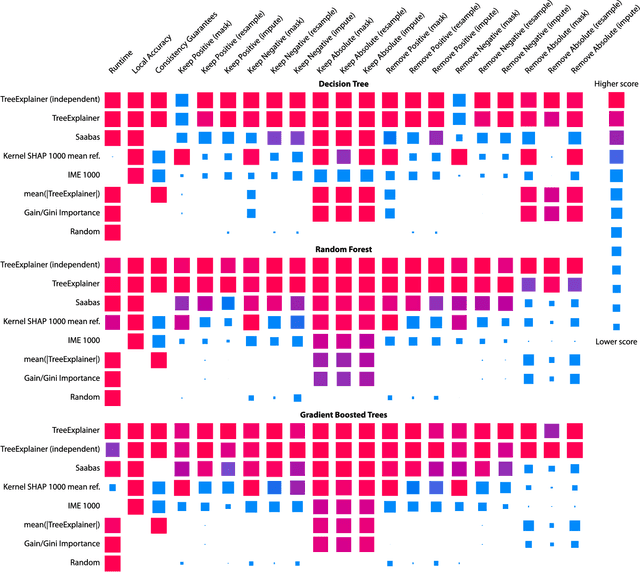
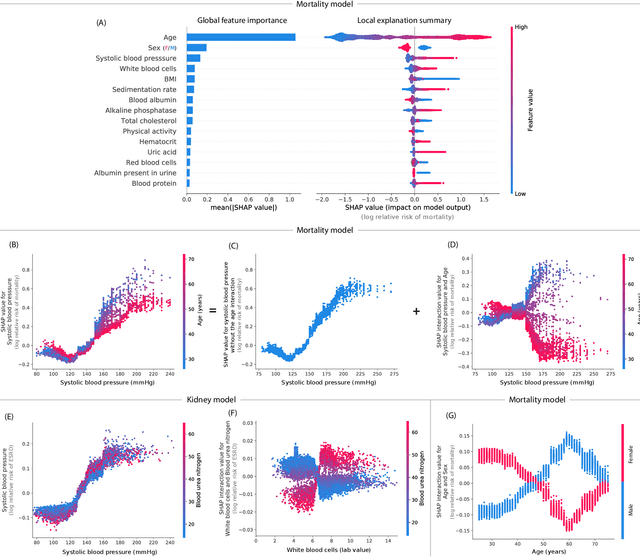
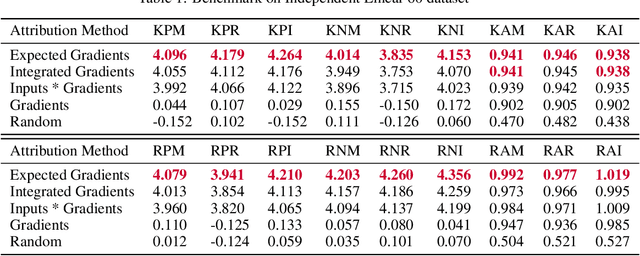
Abstract:Tree-based machine learning models such as random forests, decision trees, and gradient boosted trees are the most popular non-linear predictive models used in practice today, yet comparatively little attention has been paid to explaining their predictions. Here we significantly improve the interpretability of tree-based models through three main contributions: 1) The first polynomial time algorithm to compute optimal explanations based on game theory. 2) A new type of explanation that directly measures local feature interaction effects. 3) A new set of tools for understanding global model structure based on combining many local explanations of each prediction. We apply these tools to three medical machine learning problems and show how combining many high-quality local explanations allows us to represent global structure while retaining local faithfulness to the original model. These tools enable us to i) identify high magnitude but low frequency non-linear mortality risk factors in the general US population, ii) highlight distinct population sub-groups with shared risk characteristics, iii) identify non-linear interaction effects among risk factors for chronic kidney disease, and iv) monitor a machine learning model deployed in a hospital by identifying which features are degrading the model's performance over time. Given the popularity of tree-based machine learning models, these improvements to their interpretability have implications across a broad set of domains.
Anesthesiologist-level forecasting of hypoxemia with only SpO2 data using deep learning
Dec 02, 2017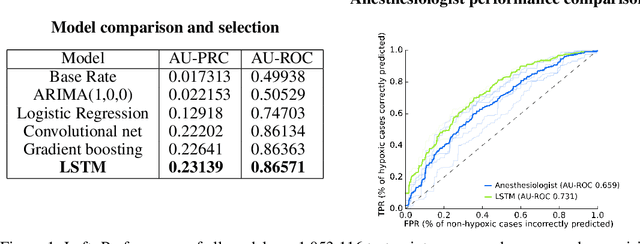
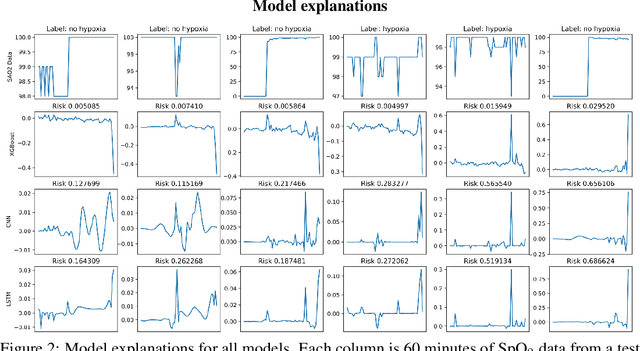
Abstract:We use a deep learning model trained only on a patient's blood oxygenation data (measurable with an inexpensive fingertip sensor) to predict impending hypoxemia (low blood oxygen) more accurately than trained anesthesiologists with access to all the data recorded in a modern operating room. We also provide a simple way to visualize the reason why a patient's risk is low or high by assigning weight to the patient's past blood oxygen values. This work has the potential to provide cutting-edge clinical decision support in low-resource settings, where rates of surgical complication and death are substantially greater than in high-resource areas.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge