Joseph D. Janizek
Visual concept ranking uncovers medical shortcuts used by large multimodal models
Feb 04, 2026Abstract:Ensuring the reliability of machine learning models in safety-critical domains such as healthcare requires auditing methods that can uncover model shortcomings. We introduce a method for identifying important visual concepts within large multimodal models (LMMs) and use it to investigate the behaviors these models exhibit when prompted with medical tasks. We primarily focus on the task of classifying malignant skin lesions from clinical dermatology images, with supplemental experiments including both chest radiographs and natural images. After showing how LMMs display unexpected gaps in performance between different demographic subgroups when prompted with demonstrating examples, we apply our method, Visual Concept Ranking (VCR), to these models and prompts. VCR generates hypotheses related to different visual feature dependencies, which we are then able to validate with manual interventions.
LAB-Bench: Measuring Capabilities of Language Models for Biology Research
Jul 16, 2024Abstract:There is widespread optimism that frontier Large Language Models (LLMs) and LLM-augmented systems have the potential to rapidly accelerate scientific discovery across disciplines. Today, many benchmarks exist to measure LLM knowledge and reasoning on textbook-style science questions, but few if any benchmarks are designed to evaluate language model performance on practical tasks required for scientific research, such as literature search, protocol planning, and data analysis. As a step toward building such benchmarks, we introduce the Language Agent Biology Benchmark (LAB-Bench), a broad dataset of over 2,400 multiple choice questions for evaluating AI systems on a range of practical biology research capabilities, including recall and reasoning over literature, interpretation of figures, access and navigation of databases, and comprehension and manipulation of DNA and protein sequences. Importantly, in contrast to previous scientific benchmarks, we expect that an AI system that can achieve consistently high scores on the more difficult LAB-Bench tasks would serve as a useful assistant for researchers in areas such as literature search and molecular cloning. As an initial assessment of the emergent scientific task capabilities of frontier language models, we measure performance of several against our benchmark and report results compared to human expert biology researchers. We will continue to update and expand LAB-Bench over time, and expect it to serve as a useful tool in the development of automated research systems going forward. A public subset of LAB-Bench is available for use at the following URL: https://huggingface.co/datasets/futurehouse/lab-bench
True to the Model or True to the Data?
Jun 29, 2020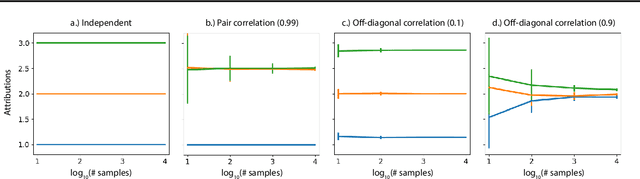
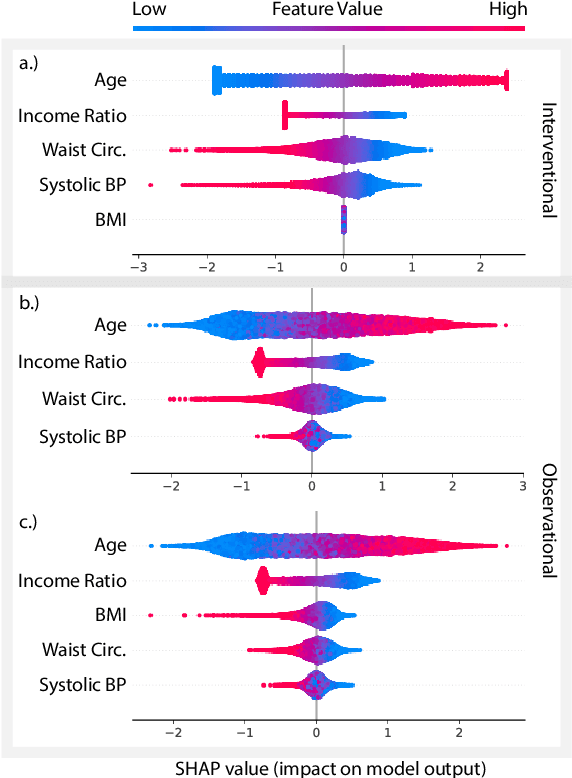
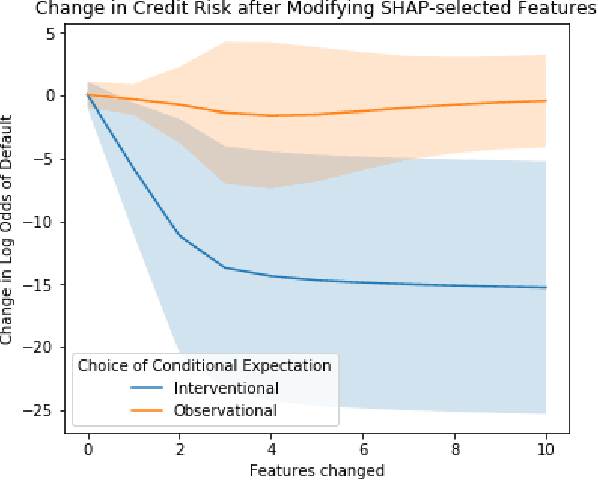
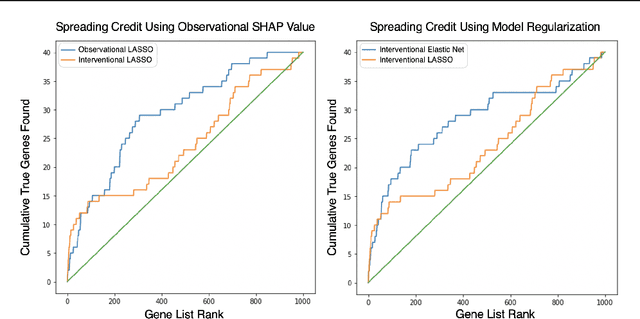
Abstract:A variety of recent papers discuss the application of Shapley values, a concept for explaining coalitional games, for feature attribution in machine learning. However, the correct way to connect a machine learning model to a coalitional game has been a source of controversy. The two main approaches that have been proposed differ in the way that they condition on known features, using either (1) an interventional or (2) an observational conditional expectation. While previous work has argued that one of the two approaches is preferable in general, we argue that the choice is application dependent. Furthermore, we argue that the choice comes down to whether it is desirable to be true to the model or true to the data. We use linear models to investigate this choice. After deriving an efficient method for calculating observational conditional expectation Shapley values for linear models, we investigate how correlation in simulated data impacts the convergence of observational conditional expectation Shapley values. Finally, we present two real data examples that we consider to be representative of possible use cases for feature attribution -- (1) credit risk modeling and (2) biological discovery. We show how a different choice of value function performs better in each scenario, and how possible attributions are impacted by modeling choices.
Explaining Explanations: Axiomatic Feature Interactions for Deep Networks
Feb 12, 2020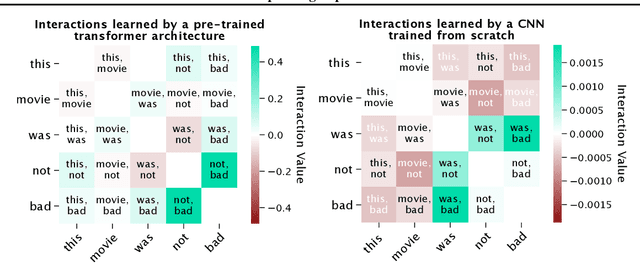
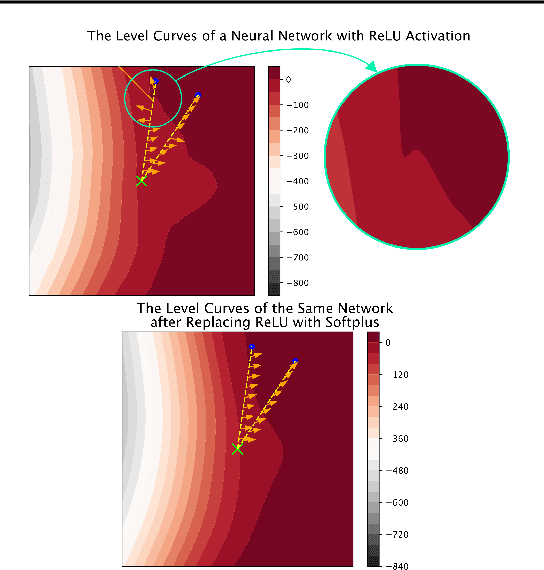

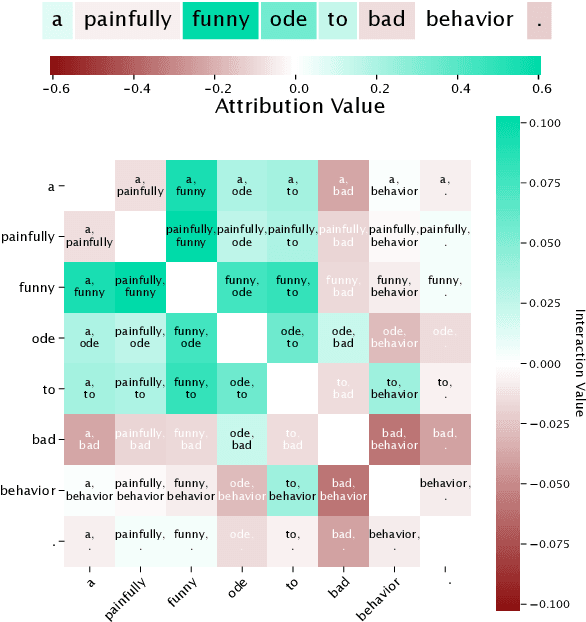
Abstract:Recent work has shown great promise in explaining neural network behavior. In particular, feature attribution methods explain which features were most important to a model's prediction on a given input. However, for many tasks, simply knowing which features were important to a model's prediction may not provide enough insight to understand model behavior. The interactions between features within the model may better help us understand not only the model, but also why certain features are more important than others. In this work we present Integrated Hessians, an extension of Integrated Gradients that explains pairwise feature interactions in neural networks. Integrated Hessians overcomes several theoretical limitations of previous methods to explain interactions, and unlike such previous methods is not limited to a specific architecture or class of neural network. We apply Integrated Hessians on a variety of neural networks trained on language data, biological data, astronomy data, and medical data and gain new insight into model behavior in each domain. Code available at https://github.com/suinleelab/path_explain
An Adversarial Approach for the Robust Classification of Pneumonia from Chest Radiographs
Jan 13, 2020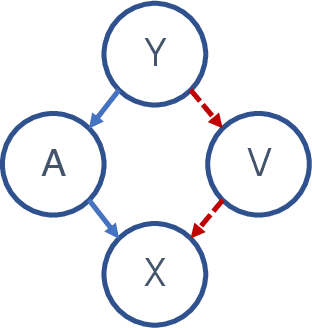
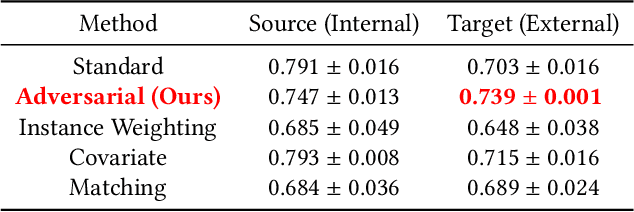
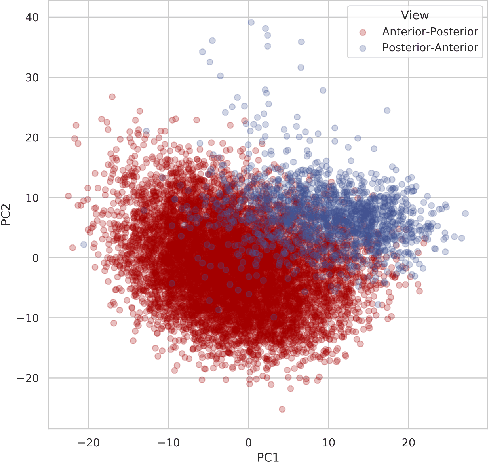
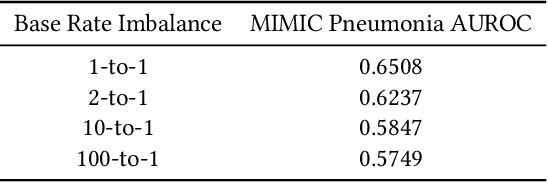
Abstract:While deep learning has shown promise in the domain of disease classification from medical images, models based on state-of-the-art convolutional neural network architectures often exhibit performance loss due to dataset shift. Models trained using data from one hospital system achieve high predictive performance when tested on data from the same hospital, but perform significantly worse when they are tested in different hospital systems. Furthermore, even within a given hospital system, deep learning models have been shown to depend on hospital- and patient-level confounders rather than meaningful pathology to make classifications. In order for these models to be safely deployed, we would like to ensure that they do not use confounding variables to make their classification, and that they will work well even when tested on images from hospitals that were not included in the training data. We attempt to address this problem in the context of pneumonia classification from chest radiographs. We propose an approach based on adversarial optimization, which allows us to learn more robust models that do not depend on confounders. Specifically, we demonstrate improved out-of-hospital generalization performance of a pneumonia classifier by training a model that is invariant to the view position of chest radiographs (anterior-posterior vs. posterior-anterior). Our approach leads to better predictive performance on external hospital data than both a standard baseline and previously proposed methods to handle confounding, and also suggests a method for identifying models that may rely on confounders. Code available at https://github.com/suinleelab/cxr_adv.
Learning Explainable Models Using Attribution Priors
Jun 25, 2019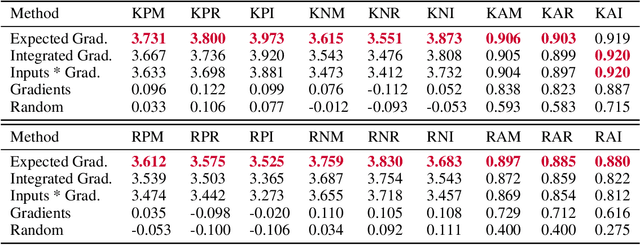

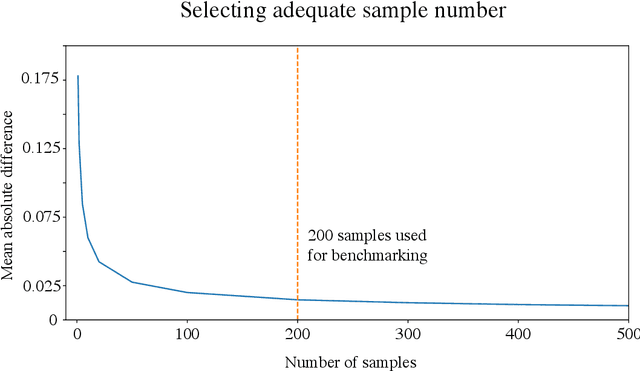
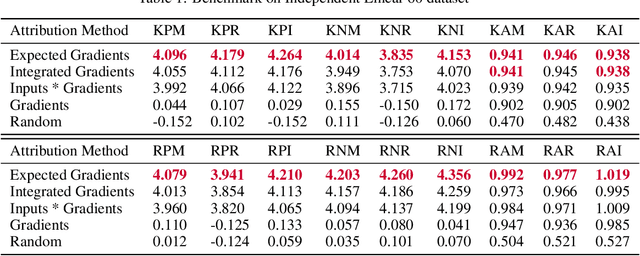
Abstract:Two important topics in deep learning both involve incorporating humans into the modeling process: Model priors transfer information from humans to a model by constraining the model's parameters; Model attributions transfer information from a model to humans by explaining the model's behavior. We propose connecting these topics with attribution priors (https://github.com/suinleelab/attributionpriors), which allow humans to use the common language of attributions to enforce prior expectations about a model's behavior during training. We develop a differentiable axiomatic feature attribution method called expected gradients and show how to directly regularize these attributions during training. We demonstrate the broad applicability of attribution priors ($\Omega$) by presenting three distinct examples that regularize models to behave more intuitively in three different domains: 1) on image data, $\Omega_{\textrm{pixel}}$ encourages models to have piecewise smooth attribution maps; 2) on gene expression data, $\Omega_{\textrm{graph}}$ encourages models to treat functionally related genes similarly; 3) on a health care dataset, $\Omega_{\textrm{sparse}}$ encourages models to rely on fewer features. In all three domains, attribution priors produce models with more intuitive behavior and better generalization performance by encoding constraints that would otherwise be very difficult to encode using standard model priors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge