Hugh Chen
Contrastive Corpus Attribution for Explaining Representations
Sep 30, 2022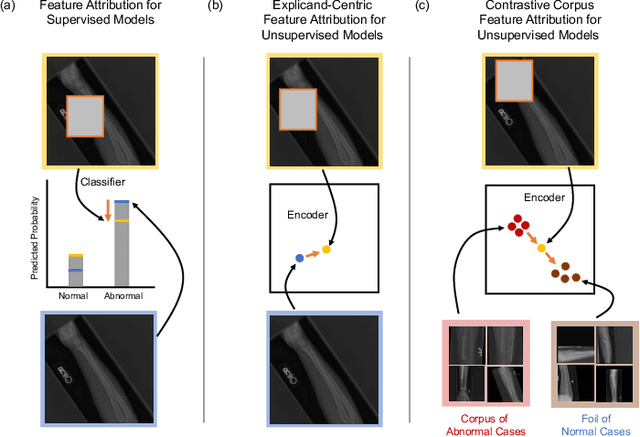
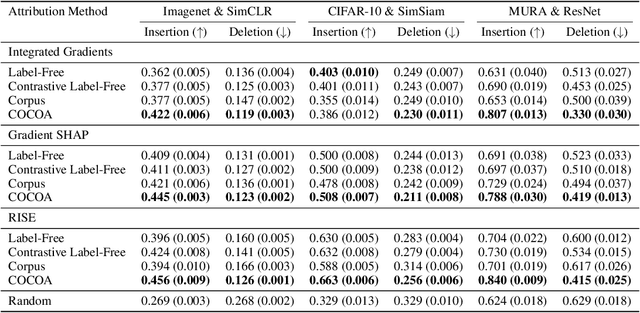
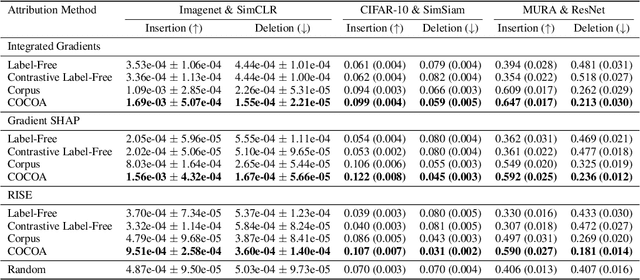
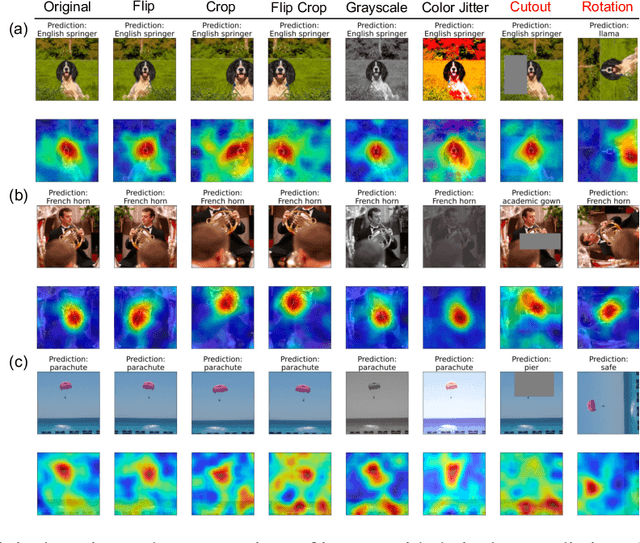
Abstract:Despite the widespread use of unsupervised models, very few methods are designed to explain them. Most explanation methods explain a scalar model output. However, unsupervised models output representation vectors, the elements of which are not good candidates to explain because they lack semantic meaning. To bridge this gap, recent works defined a scalar explanation output: a dot product-based similarity in the representation space to the sample being explained (i.e., an explicand). Although this enabled explanations of unsupervised models, the interpretation of this approach can still be opaque because similarity to the explicand's representation may not be meaningful to humans. To address this, we propose contrastive corpus similarity, a novel and semantically meaningful scalar explanation output based on a reference corpus and a contrasting foil set of samples. We demonstrate that contrastive corpus similarity is compatible with many post-hoc feature attribution methods to generate COntrastive COrpus Attributions (COCOA) and quantitatively verify that features important to the corpus are identified. We showcase the utility of COCOA in two ways: (i) we draw insights by explaining augmentations of the same image in a contrastive learning setting (SimCLR); and (ii) we perform zero-shot object localization by explaining the similarity of image representations to jointly learned text representations (CLIP).
Algorithms to estimate Shapley value feature attributions
Jul 15, 2022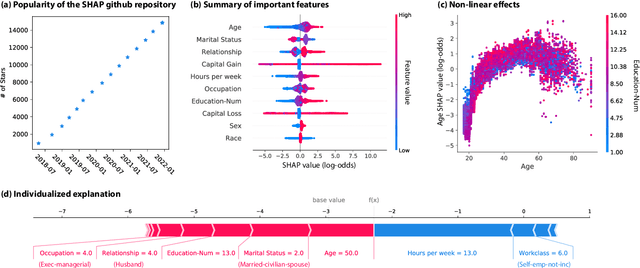
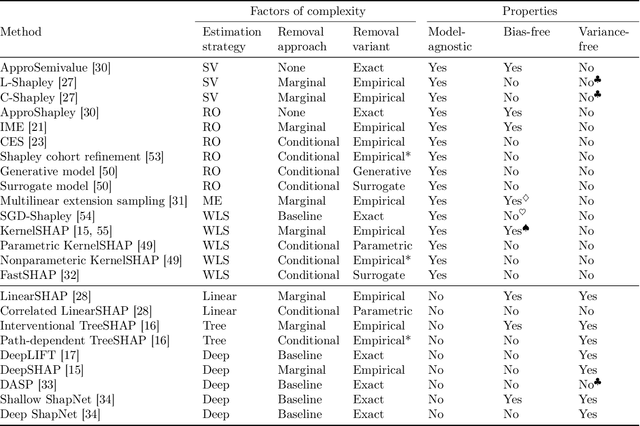
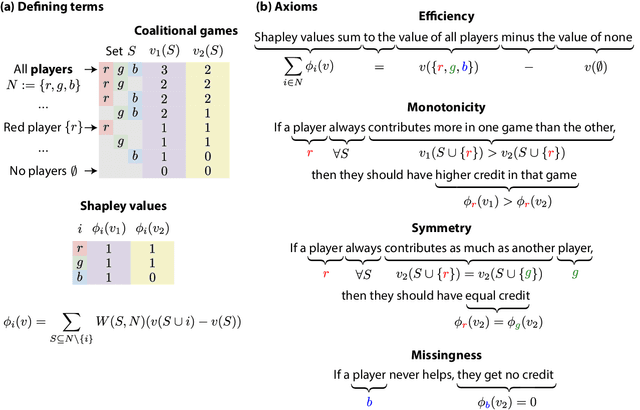
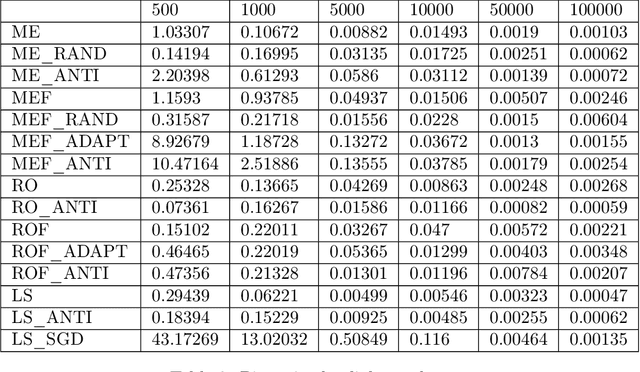
Abstract:Feature attributions based on the Shapley value are popular for explaining machine learning models; however, their estimation is complex from both a theoretical and computational standpoint. We disentangle this complexity into two factors: (1)~the approach to removing feature information, and (2)~the tractable estimation strategy. These two factors provide a natural lens through which we can better understand and compare 24 distinct algorithms. Based on the various feature removal approaches, we describe the multiple types of Shapley value feature attributions and methods to calculate each one. Then, based on the tractable estimation strategies, we characterize two distinct families of approaches: model-agnostic and model-specific approximations. For the model-agnostic approximations, we benchmark a wide class of estimation approaches and tie them to alternative yet equivalent characterizations of the Shapley value. For the model-specific approximations, we clarify the assumptions crucial to each method's tractability for linear, tree, and deep models. Finally, we identify gaps in the literature and promising future research directions.
Explaining a Series of Models by Propagating Local Feature Attributions
Apr 30, 2021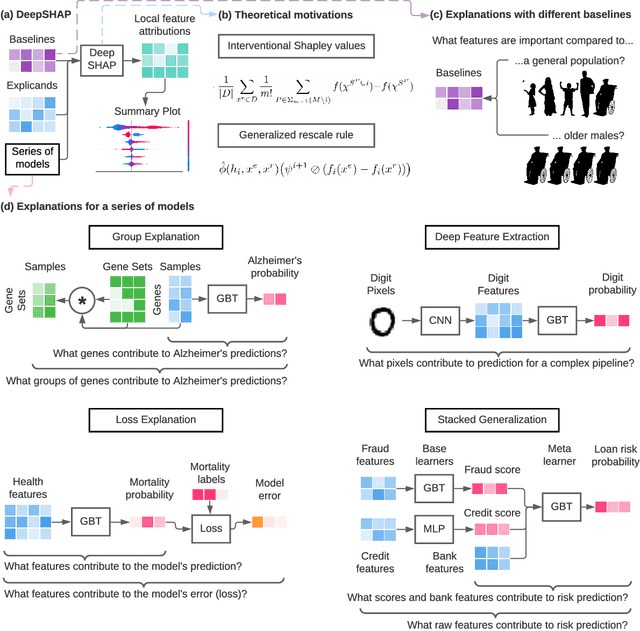

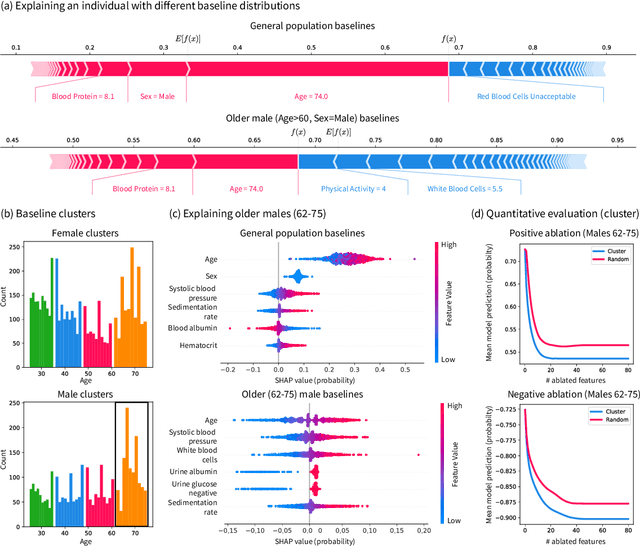
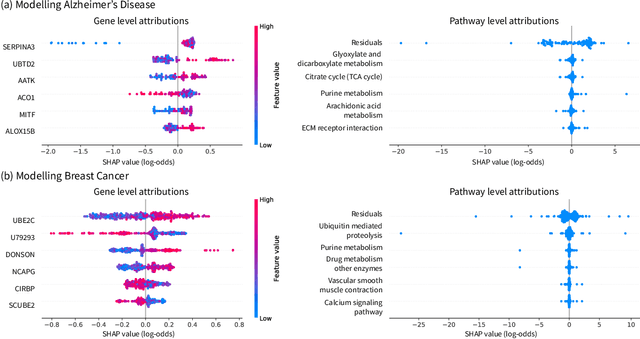
Abstract:Pipelines involving a series of several machine learning models (e.g., stacked generalization ensembles, neural network feature extractors) improve performance in many domains but are difficult to understand. To improve their transparency, we introduce a framework to propagate local feature attributions through complex pipelines of models based on a connection to the Shapley value. Our framework enables us to (1) draw higher-level conclusions based on groups of gene expression features for Alzheimer's and breast cancer histologic grade prediction, (2) draw important insights about the errors a mortality prediction model makes by explaining a loss that is a non-linear transformation of the model's output, (3) explain pipelines of deep feature extractors fed into a tree model for MNIST digit classification, and (4) interpret important consumer scores and raw features in a stacked generalization setting to predict risk for home equity line of credit applications. Importantly, in the consumer scoring example, DeepSHAP is the only feature attribution technique we are aware of that allows independent entities (e.g., lending institutions, credit bureaus) to compute attributions for the original features without having to share their proprietary models. Quantitatively comparing our framework to model-agnostic approaches, we show that our approach is an order of magnitude faster while providing equally salient explanations. In addition, we describe how to incorporate an empirical baseline distribution, which allows us to (1) demonstrate the bias of previous approaches that use a single baseline sample, and (2) present a straightforward methodology for choosing meaningful baseline distributions.
True to the Model or True to the Data?
Jun 29, 2020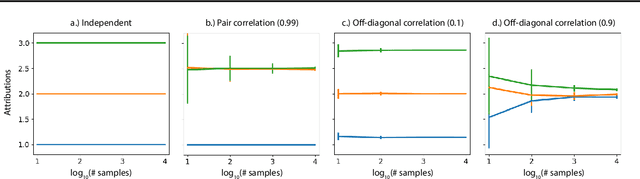
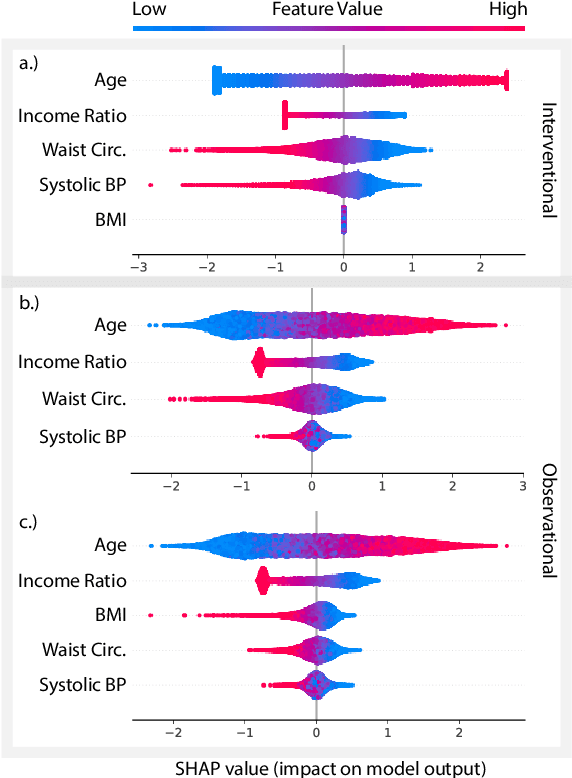
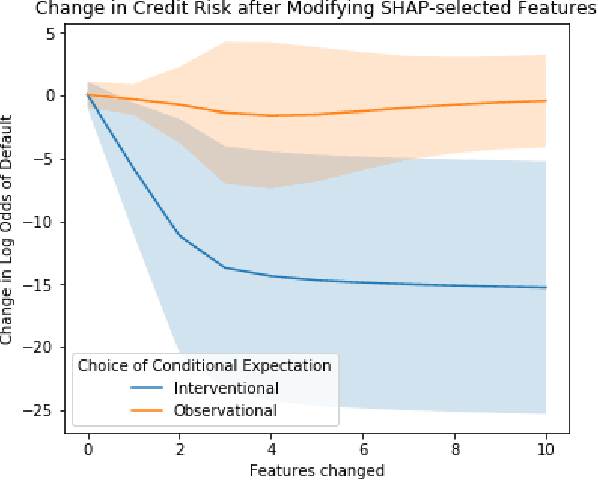
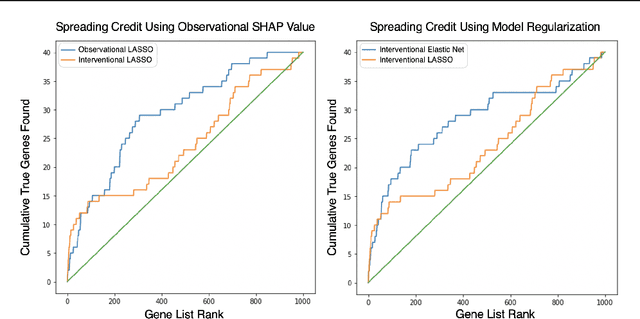
Abstract:A variety of recent papers discuss the application of Shapley values, a concept for explaining coalitional games, for feature attribution in machine learning. However, the correct way to connect a machine learning model to a coalitional game has been a source of controversy. The two main approaches that have been proposed differ in the way that they condition on known features, using either (1) an interventional or (2) an observational conditional expectation. While previous work has argued that one of the two approaches is preferable in general, we argue that the choice is application dependent. Furthermore, we argue that the choice comes down to whether it is desirable to be true to the model or true to the data. We use linear models to investigate this choice. After deriving an efficient method for calculating observational conditional expectation Shapley values for linear models, we investigate how correlation in simulated data impacts the convergence of observational conditional expectation Shapley values. Finally, we present two real data examples that we consider to be representative of possible use cases for feature attribution -- (1) credit risk modeling and (2) biological discovery. We show how a different choice of value function performs better in each scenario, and how possible attributions are impacted by modeling choices.
Deep Transfer Learning for Physiological Signals
Feb 12, 2020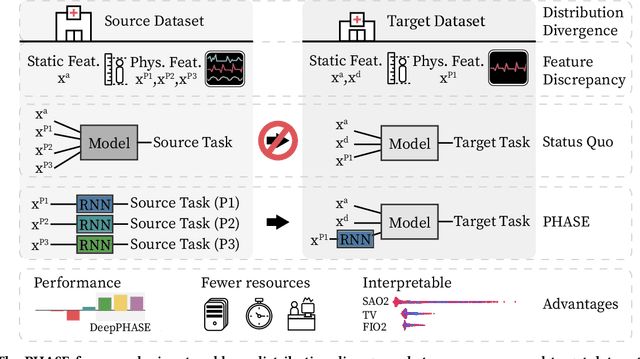
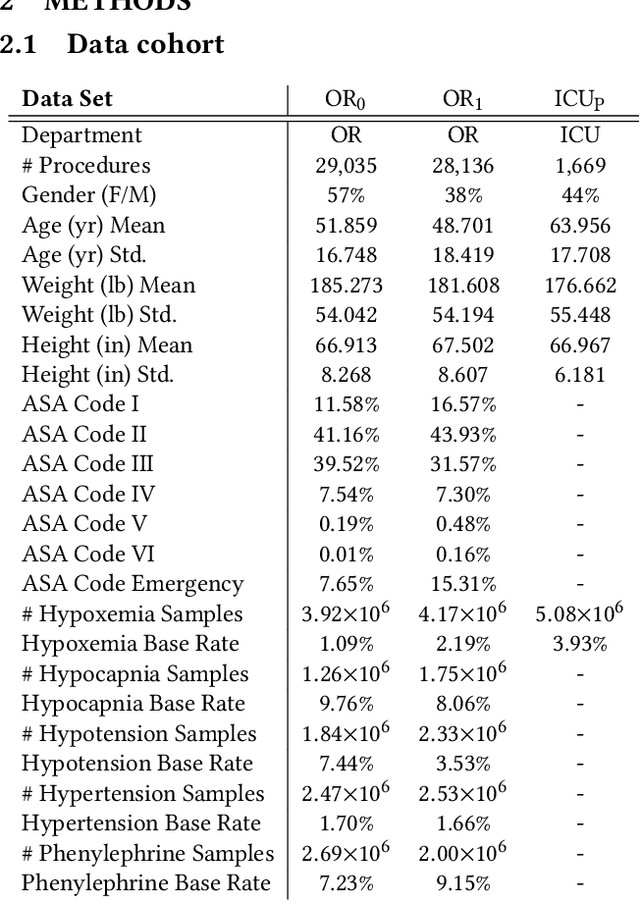
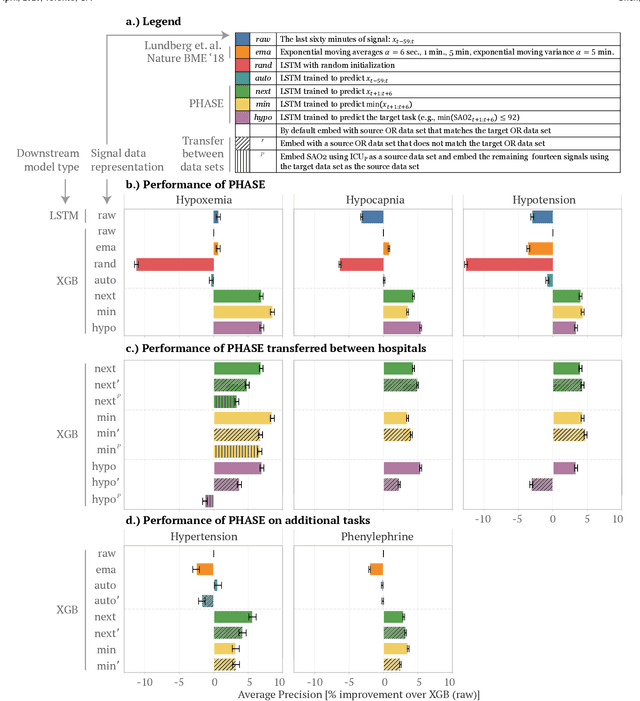
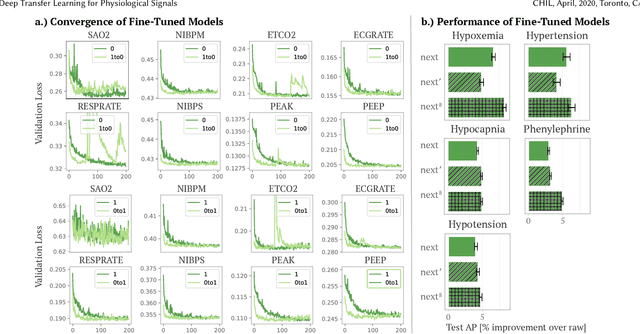
Abstract:Deep learning is increasingly common in healthcare, yet transfer learning for physiological signals (e.g., temperature, heart rate, etc.) is under-explored. Here, we present a straightforward, yet performant framework for transferring knowledge about physiological signals. Our framework is called PHASE (PHysiologicAl Signal Embeddings). It i) learns deep embeddings of physiological signals and ii) predicts adverse outcomes based on the embeddings. PHASE is the first instance of deep transfer learning in a cross-hospital, cross-department setting for physiological signals. We show that PHASE's per-signal (one for each signal) LSTM embedding functions confer a number of benefits including improved performance, successful transference between hospitals, and lower computational cost.
Explaining Models by Propagating Shapley Values of Local Components
Nov 27, 2019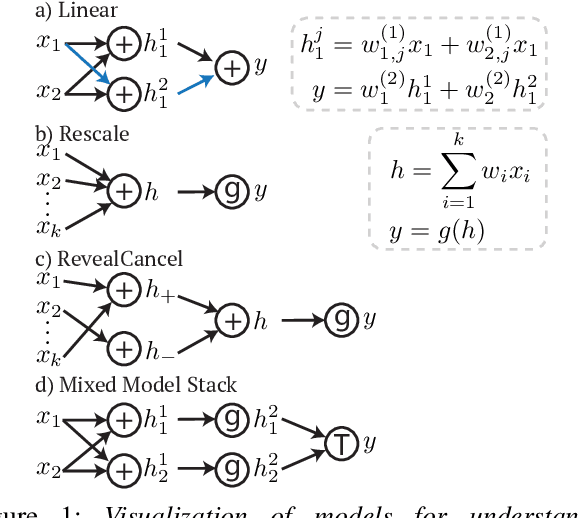


Abstract:In healthcare, making the best possible predictions with complex models (e.g., neural networks, ensembles/stacks of different models) can impact patient welfare. In order to make these complex models explainable, we present DeepSHAP for mixed model types, a framework for layer wise propagation of Shapley values that builds upon DeepLIFT (an existing approach for explaining neural networks). We show that in addition to being able to explain neural networks, this new framework naturally enables attributions for stacks of mixed models (e.g., neural network feature extractor into a tree model) as well as attributions of the loss. Finally, we theoretically justify a method for obtaining attributions with respect to a background distribution (under a Shapley value framework).
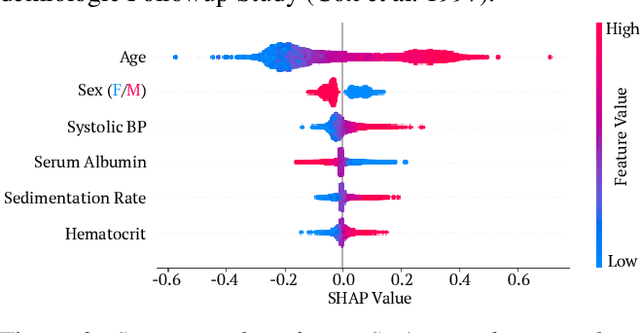
Explainable AI for Trees: From Local Explanations to Global Understanding
May 11, 2019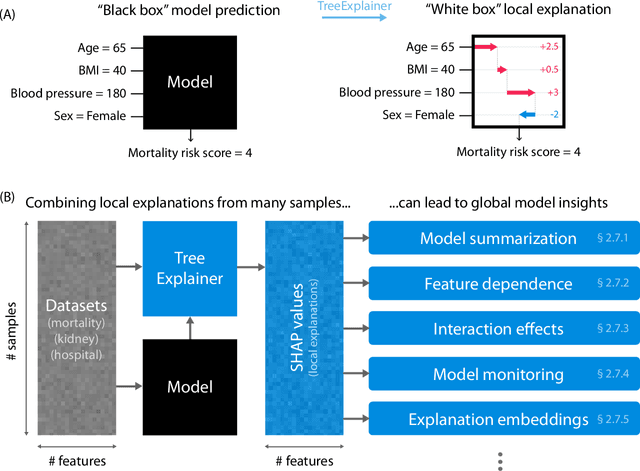
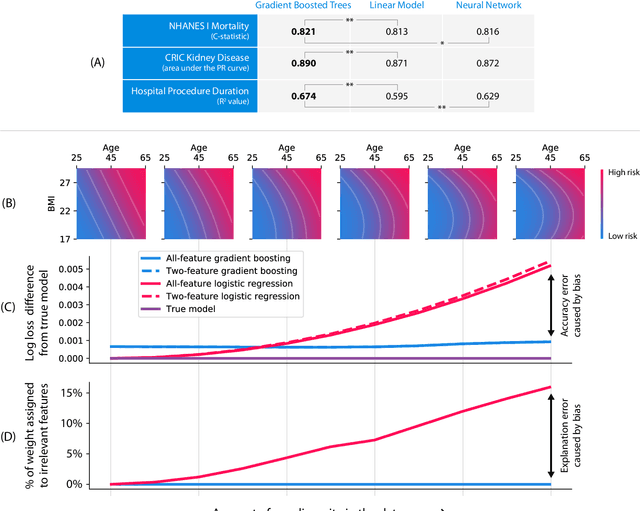
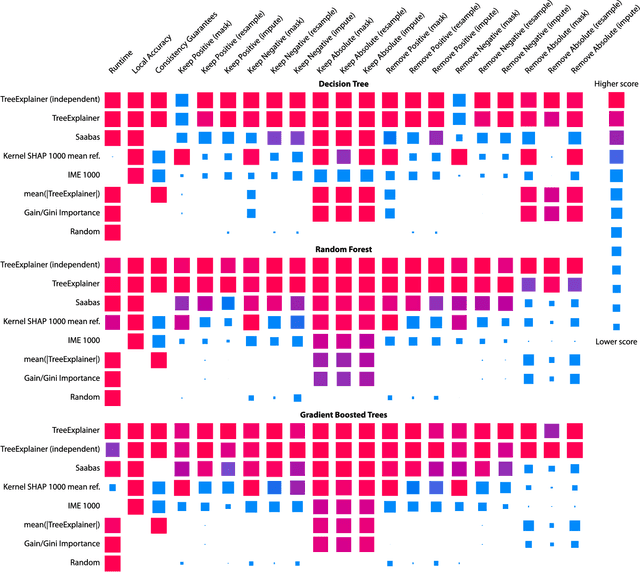
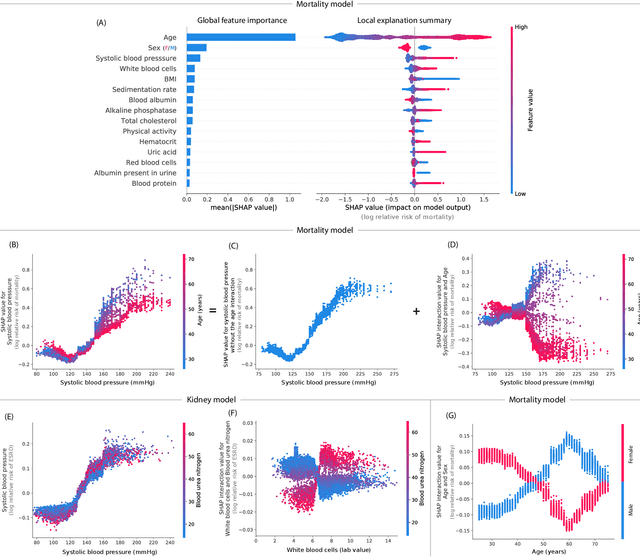
Abstract:Tree-based machine learning models such as random forests, decision trees, and gradient boosted trees are the most popular non-linear predictive models used in practice today, yet comparatively little attention has been paid to explaining their predictions. Here we significantly improve the interpretability of tree-based models through three main contributions: 1) The first polynomial time algorithm to compute optimal explanations based on game theory. 2) A new type of explanation that directly measures local feature interaction effects. 3) A new set of tools for understanding global model structure based on combining many local explanations of each prediction. We apply these tools to three medical machine learning problems and show how combining many high-quality local explanations allows us to represent global structure while retaining local faithfulness to the original model. These tools enable us to i) identify high magnitude but low frequency non-linear mortality risk factors in the general US population, ii) highlight distinct population sub-groups with shared risk characteristics, iii) identify non-linear interaction effects among risk factors for chronic kidney disease, and iv) monitor a machine learning model deployed in a hospital by identifying which features are degrading the model's performance over time. Given the popularity of tree-based machine learning models, these improvements to their interpretability have implications across a broad set of domains.
Hybrid Gradient Boosting Trees and Neural Networks for Forecasting Operating Room Data
Jan 24, 2018



Abstract:Time series data constitutes a distinct and growing problem in machine learning. As the corpus of time series data grows larger, deep models that simultaneously learn features and classify with these features can be intractable or suboptimal. In this paper, we present feature learning via long short term memory (LSTM) networks and prediction via gradient boosting trees (XGB). Focusing on the consequential setting of electronic health record data, we predict the occurrence of hypoxemia five minutes into the future based on past features. We make two observations: 1) long short term memory networks are effective at capturing long term dependencies based on a single feature and 2) gradient boosting trees are capable of tractably combining a large number of features including static features like height and weight. With these observations in mind, we generate features by performing "supervised" representation learning with LSTM networks. Augmenting the original XGB model with these features gives significantly better performance than either individual method.
Anesthesiologist-level forecasting of hypoxemia with only SpO2 data using deep learning
Dec 02, 2017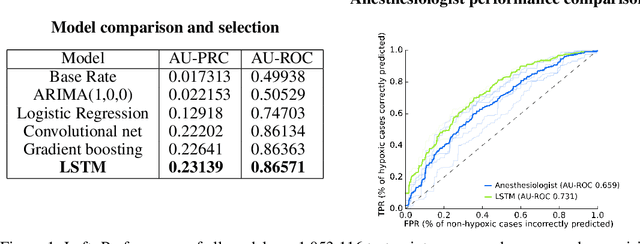
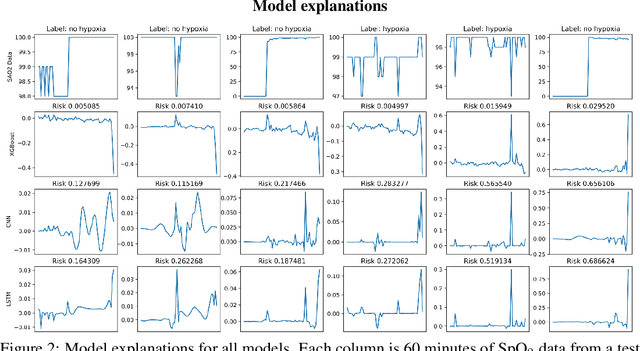
Abstract:We use a deep learning model trained only on a patient's blood oxygenation data (measurable with an inexpensive fingertip sensor) to predict impending hypoxemia (low blood oxygen) more accurately than trained anesthesiologists with access to all the data recorded in a modern operating room. We also provide a simple way to visualize the reason why a patient's risk is low or high by assigning weight to the patient's past blood oxygen values. This work has the potential to provide cutting-edge clinical decision support in low-resource settings, where rates of surgical complication and death are substantially greater than in high-resource areas.
Checkpoint Ensembles: Ensemble Methods from a Single Training Process
Oct 09, 2017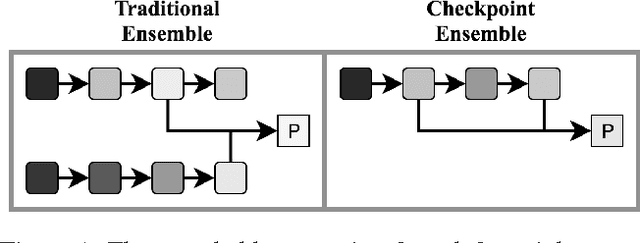



Abstract:We present the checkpoint ensembles method that can learn ensemble models on a single training process. Although checkpoint ensembles can be applied to any parametric iterative learning technique, here we focus on neural networks. Neural networks' composable and simple neurons make it possible to capture many individual and interaction effects among features. However, small sample sizes and sampling noise may result in patterns in the training data that are not representative of the true relationship between the features and the outcome. As a solution, regularization during training is often used (e.g. dropout). However, regularization is no panacea -- it does not perfectly address overfitting. Even with methods like dropout, two methodologies are commonly used in practice. First is to utilize a validation set independent to the training set as a way to decide when to stop training. Second is to use ensemble methods to further reduce overfitting and take advantage of local optima (i.e. averaging over the predictions of several models). In this paper, we explore checkpoint ensembles -- a simple technique that combines these two ideas in one training process. Checkpoint ensembles improve performance by averaging the predictions from "checkpoints" of the best models within single training process. We use three real-world data sets -- text, image, and electronic health record data -- using three prediction models: a vanilla neural network, a convolutional neural network, and a long short term memory network to show that checkpoint ensembles outperform existing methods: a method that selects a model by minimum validation score, and two methods that average models by weights. Our results also show that checkpoint ensembles capture a portion of the performance gains that traditional ensembles provide.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge