Saige Rutherford
Self-Supervised Masked Mesh Learning for Unsupervised Anomaly Detection on 3D Cortical Surfaces
Dec 07, 2024


Abstract:Unsupervised anomaly detection in brain imaging is challenging. In this paper, we propose a self-supervised masked mesh learning for unsupervised anomaly detection in 3D cortical surfaces. Our framework leverages the intrinsic geometry of the cortical surface to learn a self-supervised representation that captures the underlying structure of the brain. We introduce a masked mesh convolutional neural network (MMN) that learns to predict masked regions of the cortical surface. By training the MMN on a large dataset of healthy subjects, we learn a representation that captures the normal variation in the cortical surface. We then use this representation to detect anomalies in unseen individuals by calculating anomaly scores based on the reconstruction error of the MMN. We evaluate our framework by training on population-scale dataset UKB and HCP-Aging and testing on two datasets of Alzheimer's disease patients ADNI and OASIS3. Our results show that our framework can detect anomalies in cortical thickness, cortical volume, and cortical sulcus features, which are known to be sensitive biomarkers for Alzheimer's disease. Our proposed framework provides a promising approach for unsupervised anomaly detection based on normative variation of cortical features.
To which reference class do you belong? Measuring racial fairness of reference classes with normative modeling
Jul 26, 2024



Abstract:Reference classes in healthcare establish healthy norms, such as pediatric growth charts of height and weight, and are used to chart deviations from these norms which represent potential clinical risk. How the demographics of the reference class influence clinical interpretation of deviations is unknown. Using normative modeling, a method for building reference classes, we evaluate the fairness (racial bias) in reference models of structural brain images that are widely used in psychiatry and neurology. We test whether including race in the model creates fairer models. We predict self-reported race using the deviation scores from three different reference class normative models, to better understand bias in an integrated, multivariate sense. Across all of these tasks, we uncover racial disparities that are not easily addressed with existing data or commonly used modeling techniques. Our work suggests that deviations from the norm could be due to demographic mismatch with the reference class, and assigning clinical meaning to these deviations should be done with caution. Our approach also suggests that acquiring more representative samples is an urgent research priority.
A Domain Guided CNN Architecture for Predicting Age from Structural Brain Images
Aug 11, 2018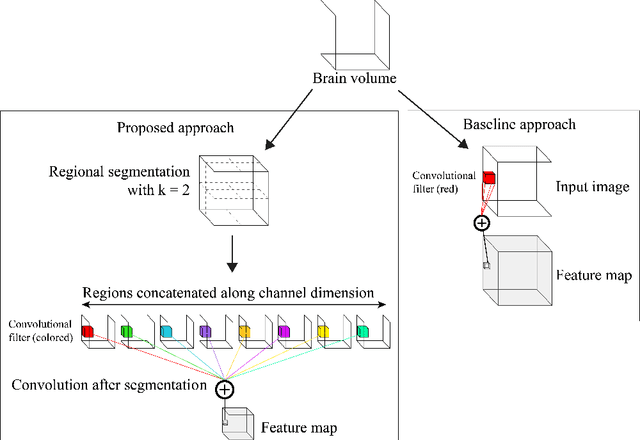
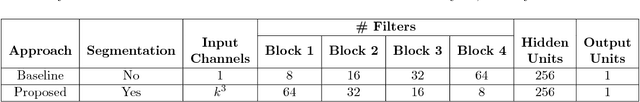
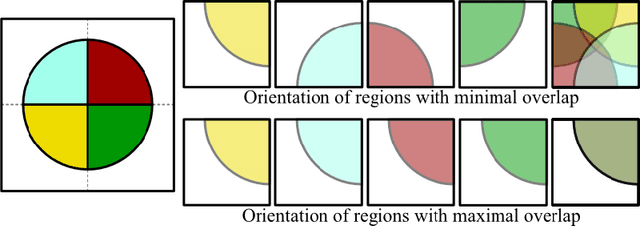
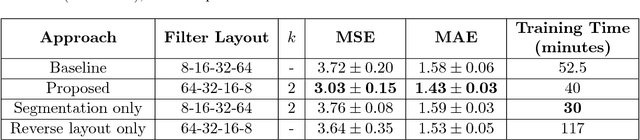
Abstract:Given the wide success of convolutional neural networks (CNNs) applied to natural images, researchers have begun to apply them to neuroimaging data. To date, however, exploration of novel CNN architectures tailored to neuroimaging data has been limited. Several recent works fail to leverage the 3D structure of the brain, instead treating the brain as a set of independent 2D slices. Approaches that do utilize 3D convolutions rely on architectures developed for object recognition tasks in natural 2D images. Such architectures make assumptions about the input that may not hold for neuroimaging. For example, existing architectures assume that patterns in the brain exhibit translation invariance. However, a pattern in the brain may have different meaning depending on where in the brain it is located. There is a need to explore novel architectures that are tailored to brain images. We present two simple modifications to existing CNN architectures based on brain image structure. Applied to the task of brain age prediction, our network achieves a mean absolute error (MAE) of 1.4 years and trains 30% faster than a CNN baseline that achieves a MAE of 1.6 years. Our results suggest that lessons learned from developing models on natural images may not directly transfer to neuroimaging tasks. Instead, there remains a large space of unexplored questions regarding model development in this area, whose answers may differ from conventional wisdom.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge