Markus Wenzel
Synthetic Generation of Dermatoscopic Images with GAN and Closed-Form Factorization
Oct 07, 2024



Abstract:In the realm of dermatological diagnoses, where the analysis of dermatoscopic and microscopic skin lesion images is pivotal for the accurate and early detection of various medical conditions, the costs associated with creating diverse and high-quality annotated datasets have hampered the accuracy and generalizability of machine learning models. We propose an innovative unsupervised augmentation solution that harnesses Generative Adversarial Network (GAN) based models and associated techniques over their latent space to generate controlled semiautomatically-discovered semantic variations in dermatoscopic images. We created synthetic images to incorporate the semantic variations and augmented the training data with these images. With this approach, we were able to increase the performance of machine learning models and set a new benchmark amongst non-ensemble based models in skin lesion classification on the HAM10000 dataset; and used the observed analytics and generated models for detailed studies on model explainability, affirming the effectiveness of our solution.
Insights Into the Inner Workings of Transformer Models for Protein Function Prediction
Sep 07, 2023Abstract:Motivation: We explored how explainable AI (XAI) can help to shed light into the inner workings of neural networks for protein function prediction, by extending the widely used XAI method of integrated gradients such that latent representations inside of transformer models, which were finetuned to Gene Ontology term and Enzyme Commission number prediction, can be inspected too. Results: The approach enabled us to identify amino acids in the sequences that the transformers pay particular attention to, and to show that these relevant sequence parts reflect expectations from biology and chemistry, both in the embedding layer and inside of the model, where we identified transformer heads with a statistically significant correspondence of attribution maps with ground truth sequence annotations (e.g., transmembrane regions, active sites) across many proteins. Availability and Implementation: Source code can be accessed at https://github.com/markuswenzel/xai-proteins .
Generative Adversarial Networks and Other Generative Models
Jul 08, 2022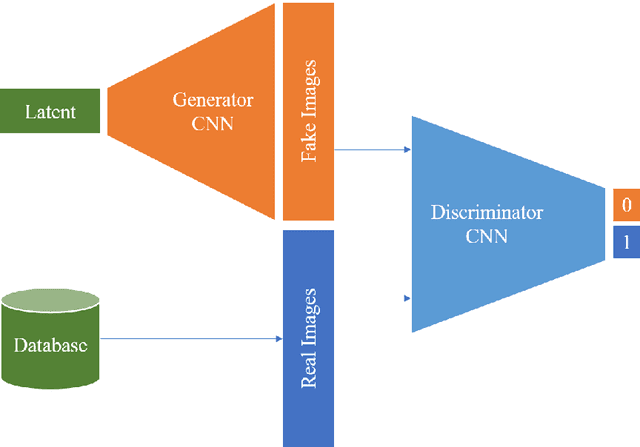
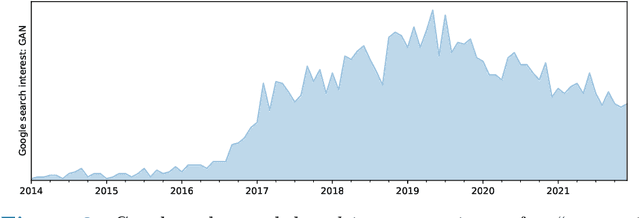
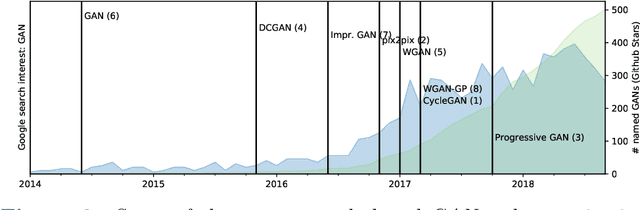
Abstract:Generative networks are fundamentally different in their aim and methods compared to CNNs for classification, segmentation, or object detection. They have initially not been meant to be an image analysis tool, but to produce naturally looking images. The adversarial training paradigm has been proposed to stabilize generative methods, and has proven to be highly successful -- though by no means from the first attempt. This chapter gives a basic introduction into the motivation for Generative Adversarial Networks (GANs) and traces the path of their success by abstracting the basic task and working mechanism, and deriving the difficulty of early practical approaches. Methods for a more stable training will be shown, and also typical signs for poor convergence and their reasons. Though this chapter focuses on GANs that are meant for image generation and image analysis, the adversarial training paradigm itself is not specific to images, and also generalizes to tasks in image analysis. Examples of architectures for image semantic segmentation and abnormality detection will be acclaimed, before contrasting GANs with further generative modeling approaches lately entering the scene. This will allow a contextualized view on the limits but also benefits of GANs.
From CNNs to Vision Transformers -- A Comprehensive Evaluation of Deep Learning Models for Histopathology
Apr 11, 2022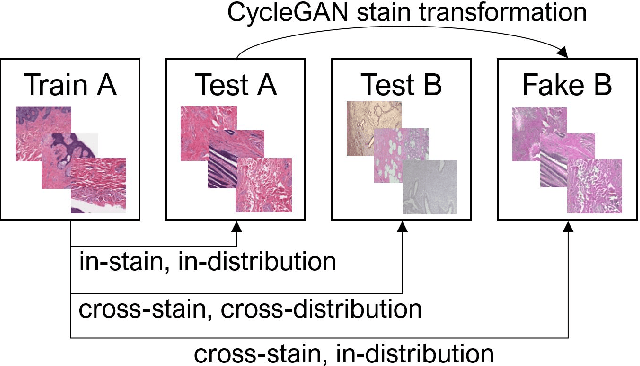



Abstract:While machine learning is currently transforming the field of histopathology, the domain lacks a comprehensive evaluation of state-of-the-art models based on essential but complementary quality requirements beyond a mere classification accuracy. In order to fill this gap, we conducted an extensive evaluation by benchmarking a wide range of classification models, including recent vision transformers, convolutional neural networks and hybrid models comprising transformer and convolutional models. We thoroughly tested the models on five widely used histopathology datasets containing whole slide images of breast, gastric, and colorectal cancer and developed a novel approach using an image-to-image translation model to assess the robustness of a cancer classification model against stain variations. Further, we extended existing interpretability methods to previously unstudied models and systematically reveal insights of the models' classification strategies that allow for plausibility checks and systematic comparisons. The study resulted in specific model recommendations for practitioners as well as putting forward a general methodology to quantify a model's quality according to complementary requirements that can be transferred to future model architectures.
Infant Brain Age Classification: 2D CNN Outperforms 3D CNN in Small Dataset
Dec 27, 2021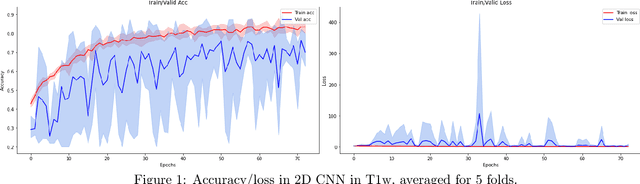
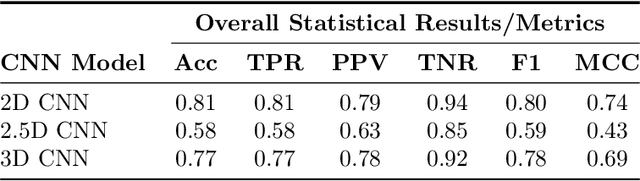
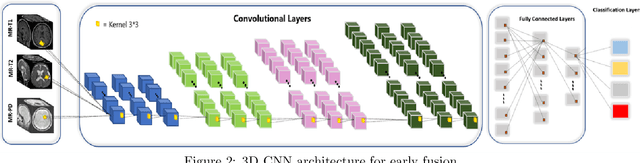
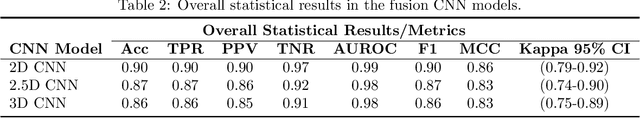
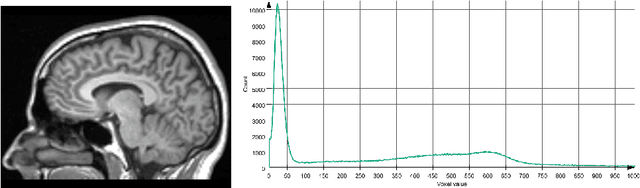
Abstract:Determining if the brain is developing normally is a key component of pediatric neuroradiology and neurology. Brain magnetic resonance imaging (MRI) of infants demonstrates a specific pattern of development beyond simply myelination. While radiologists have used myelination patterns, brain morphology and size characteristics to determine age-adequate brain maturity, this requires years of experience in pediatric neuroradiology. With no standardized criteria, visual estimation of the structural maturity of the brain from MRI before three years of age remains dominated by inter-observer and intra-observer variability. A more objective estimation of brain developmental age could help physicians identify many neurodevelopmental conditions and diseases earlier and more reliably. Such data, however, is naturally hard to obtain, and the observer ground truth not much of a gold standard due to subjectivity of assessment. In this light, we explore the general feasibility to tackle this task, and the utility of different approaches, including two- and three-dimensional convolutional neural networks (CNN) that were trained on a fusion of T1-weighted, T2-weighted, and proton density (PD) weighted sequences from 84 individual subjects divided into four age groups from birth to 3 years of age. In the best performing approach, we achieved an accuracy of 0.90 [95% CI:0.86-0.94] using a 2D CNN on a central axial thick slab. We discuss the comparison to 3D networks and show how the performance compares to the use of only one sequence (T1w). In conclusion, despite the theoretical superiority of 3D CNN approaches, in limited-data situations, such approaches are inferior to simpler architectures. The code can be found in https://github.com/shabanian2018/Age_MRI-Classification
* 8 pages, 5 figures, 3 tables. arXiv admin note: text overlap with arXiv:2010.03963
Predicting the Binding of SARS-CoV-2 Peptides to the Major Histocompatibility Complex with Recurrent Neural Networks
Apr 16, 2021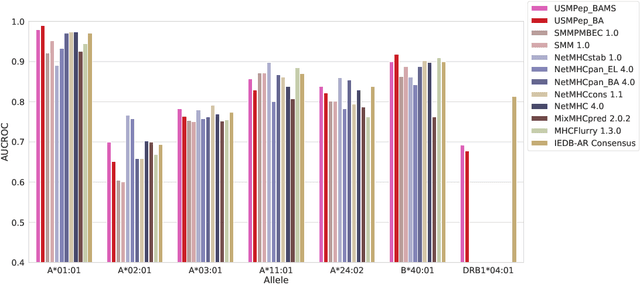
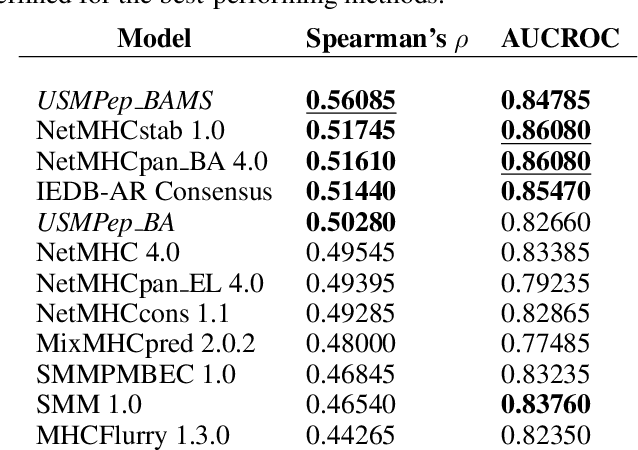
Abstract:Predicting the binding of viral peptides to the major histocompatibility complex with machine learning can potentially extend the computational immunology toolkit for vaccine development, and serve as a key component in the fight against a pandemic. In this work, we adapt and extend USMPep, a recently proposed, conceptually simple prediction algorithm based on recurrent neural networks. Most notably, we combine regressors (binding affinity data) and classifiers (mass spectrometry data) from qualitatively different data sources to obtain a more comprehensive prediction tool. We evaluate the performance on a recently released SARS-CoV-2 dataset with binding stability measurements. USMPep not only sets new benchmarks on selected single alleles, but consistently turns out to be among the best-performing methods or, for some metrics, to be even the overall best-performing method for this task.
Risk Estimation of SARS-CoV-2 Transmission from Bluetooth Low Energy Measurements
Apr 22, 2020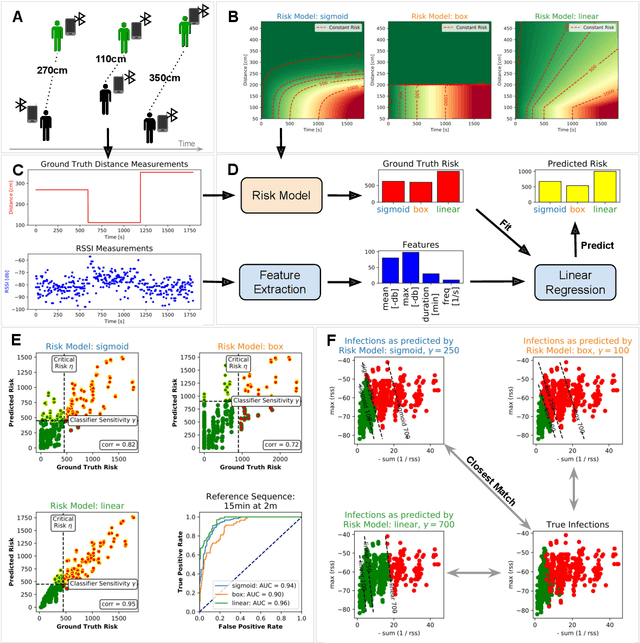
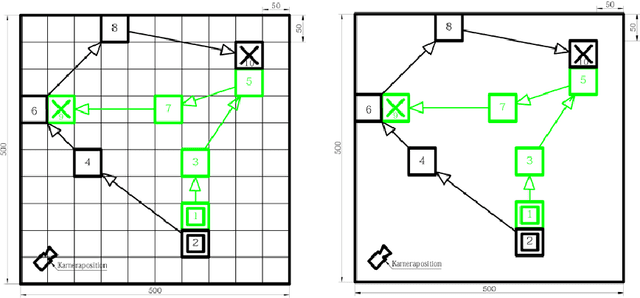
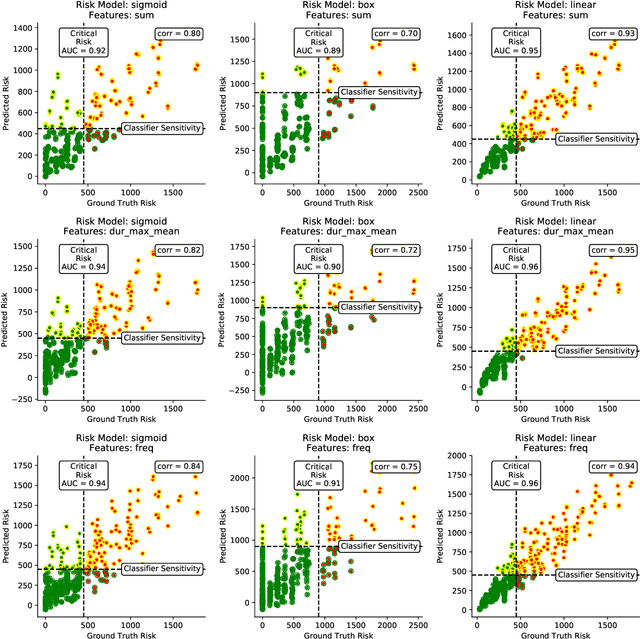
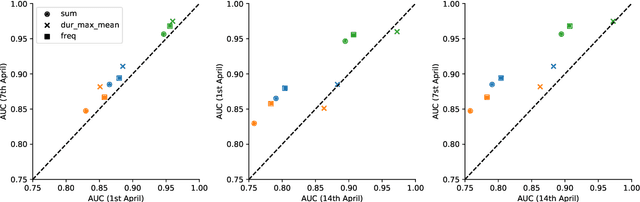
Abstract:Digital contact tracing approaches based on Bluetooth low energy (BLE) have the potential to efficiently contain and delay outbreaks of infectious diseases such as the ongoing SARS-CoV-2 pandemic. In this work we propose a novel machine learning based approach to reliably detect subjects that have spent enough time in close proximity to be at risk of being infected. Our study is an important proof of concept that will aid the battery of epidemiological policies aiming to slow down the rapid spread of COVID-19.
Focus Group on Artificial Intelligence for Health
Sep 13, 2018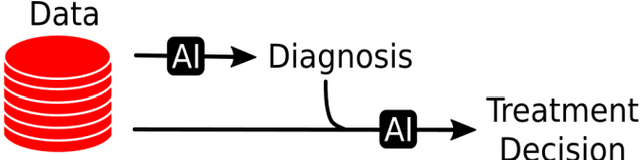
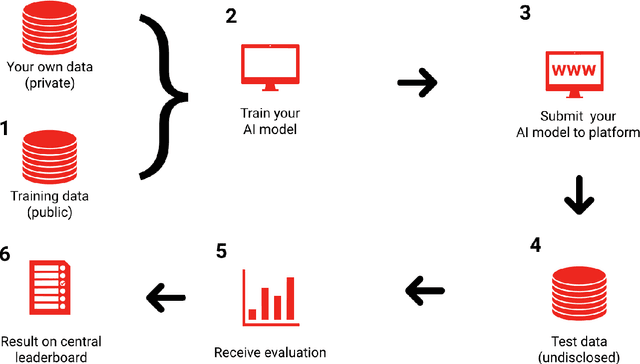
Abstract:Artificial Intelligence (AI) - the phenomenon of machines being able to solve problems that require human intelligence - has in the past decade seen an enormous rise of interest due to significant advances in effectiveness and use. The health sector, one of the most important sectors for societies and economies worldwide, is particularly interesting for AI applications, given the ongoing digitalisation of all types of health information. The potential for AI assistance in the health domain is immense, because AI can support medical decision making at reduced costs, everywhere. However, due to the complexity of AI algorithms, it is difficult to distinguish good from bad AI-based solutions and to understand their strengths and weaknesses, which is crucial for clarifying responsibilities and for building trust. For this reason, the International Telecommunication Union (ITU) has established a new Focus Group on "Artificial Intelligence for Health" (FG-AI4H) in partnership with the World Health Organization (WHO). Health and care services are usually the responsibility of a government - even when provided through private insurance systems - and thus under the responsibility of WHO/ITU member states. FG-AI4H will identify opportunities for international standardization, which will foster the application of AI to health issues on a global scale. In particular, it will establish a standardized assessment framework with open benchmarks for the evaluation of AI-based methods for health, such as AI-based diagnosis, triage or treatment decisions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge