Nils Strodthoff
ProtoMask: Segmentation-Guided Prototype Learning
Oct 01, 2025Abstract:XAI gained considerable importance in recent years. Methods based on prototypical case-based reasoning have shown a promising improvement in explainability. However, these methods typically rely on additional post-hoc saliency techniques to explain the semantics of learned prototypes. Multiple critiques have been raised about the reliability and quality of such techniques. For this reason, we study the use of prominent image segmentation foundation models to improve the truthfulness of the mapping between embedding and input space. We aim to restrict the computation area of the saliency map to a predefined semantic image patch to reduce the uncertainty of such visualizations. To perceive the information of an entire image, we use the bounding box from each generated segmentation mask to crop the image. Each mask results in an individual input in our novel model architecture named ProtoMask. We conduct experiments on three popular fine-grained classification datasets with a wide set of metrics, providing a detailed overview on explainability characteristics. The comparison with other popular models demonstrates competitive performance and unique explainability features of our model. https://github.com/uos-sis/quanproto
FeatInv: Spatially resolved mapping from feature space to input space using conditional diffusion models
May 27, 2025Abstract:Internal representations are crucial for understanding deep neural networks, such as their properties and reasoning patterns, but remain difficult to interpret. While mapping from feature space to input space aids in interpreting the former, existing approaches often rely on crude approximations. We propose using a conditional diffusion model - a pretrained high-fidelity diffusion model conditioned on spatially resolved feature maps - to learn such a mapping in a probabilistic manner. We demonstrate the feasibility of this approach across various pretrained image classifiers from CNNs to ViTs, showing excellent reconstruction capabilities. Through qualitative comparisons and robustness analysis, we validate our method and showcase possible applications, such as the visualization of concept steering in input space or investigations of the composite nature of the feature space. This approach has broad potential for improving feature space understanding in computer vision models.
Uncertainty quantification with approximate variational learning for wearable photoplethysmography prediction tasks
May 16, 2025Abstract:Photoplethysmography (PPG) signals encode information about relative changes in blood volume that can be used to assess various aspects of cardiac health non-invasively, e.g.\ to detect atrial fibrillation (AF) or predict blood pressure (BP). Deep networks are well-equipped to handle the large quantities of data acquired from wearable measurement devices. However, they lack interpretability and are prone to overfitting, leaving considerable risk for poor performance on unseen data and misdiagnosis. Here, we describe the use of two scalable uncertainty quantification techniques: Monte Carlo Dropout and the recently proposed Improved Variational Online Newton. These techniques are used to assess the trustworthiness of models trained to perform AF classification and BP regression from raw PPG time series. We find that the choice of hyperparameters has a considerable effect on the predictive performance of the models and on the quality and composition of predicted uncertainties. E.g. the stochasticity of the model parameter sampling determines the proportion of the total uncertainty that is aleatoric, and has varying effects on predictive performance and calibration quality dependent on the chosen uncertainty quantification technique and the chosen expression of uncertainty. We find significant discrepancy in the quality of uncertainties over the predicted classes, emphasising the need for a thorough evaluation protocol that assesses local and adaptive calibration. This work suggests that the choice of hyperparameters must be carefully tuned to balance predictive performance and calibration quality, and that the optimal parameterisation may vary depending on the chosen expression of uncertainty.
Machine-learning for photoplethysmography analysis: Benchmarking feature, image, and signal-based approaches
Feb 27, 2025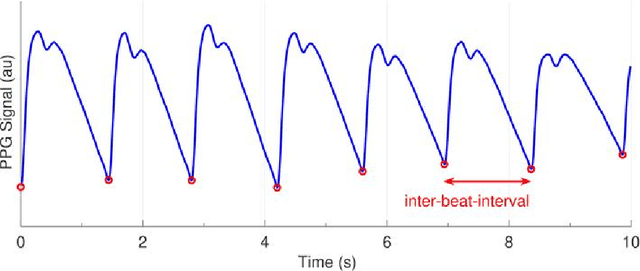
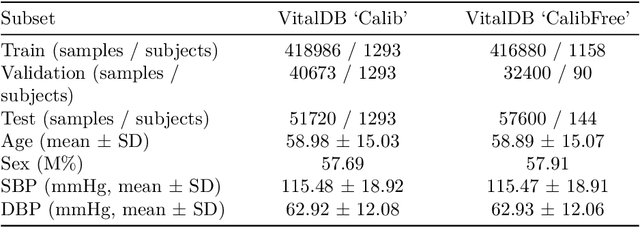
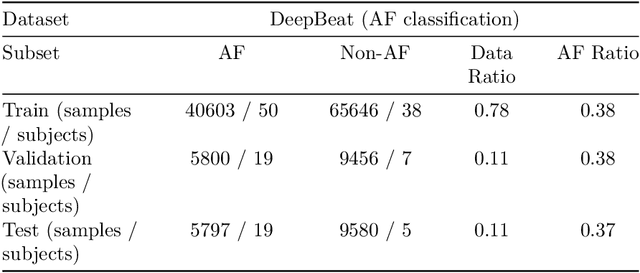
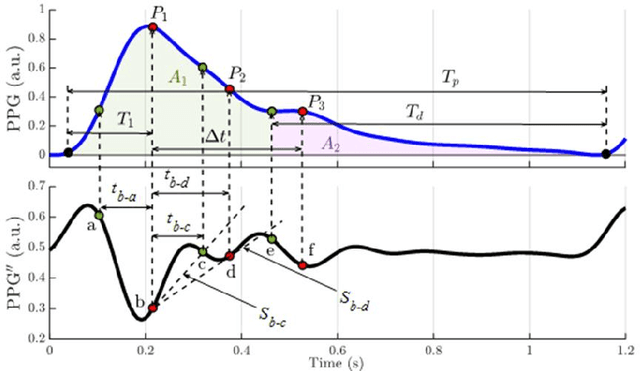
Abstract:Photoplethysmography (PPG) is a widely used non-invasive physiological sensing technique, suitable for various clinical applications. Such clinical applications are increasingly supported by machine learning methods, raising the question of the most appropriate input representation and model choice. Comprehensive comparisons, in particular across different input representations, are scarce. We address this gap in the research landscape by a comprehensive benchmarking study covering three kinds of input representations, interpretable features, image representations and raw waveforms, across prototypical regression and classification use cases: blood pressure and atrial fibrillation prediction. In both cases, the best results are achieved by deep neural networks operating on raw time series as input representations. Within this model class, best results are achieved by modern convolutional neural networks (CNNs). but depending on the task setup, shallow CNNs are often also very competitive. We envision that these results will be insightful for researchers to guide their choice on machine learning tasks for PPG data, even beyond the use cases presented in this work.
Generalizable deep learning for photoplethysmography-based blood pressure estimation -- A Benchmarking Study
Feb 26, 2025Abstract:Photoplethysmography (PPG)-based blood pressure (BP) estimation represents a promising alternative to cuff-based BP measurements. Recently, an increasing number of deep learning models have been proposed to infer BP from the raw PPG waveform. However, these models have been predominantly evaluated on in-distribution test sets, which immediately raises the question of the generalizability of these models to external datasets. To investigate this question, we trained five deep learning models on the recently released PulseDB dataset, provided in-distribution benchmarking results on this dataset, and then assessed out-of-distribution performance on several external datasets. The best model (XResNet1d101) achieved in-distribution MAEs of 9.4 and 6.0 mmHg for systolic and diastolic BP respectively on PulseDB (with subject-specific calibration), and 14.0 and 8.5 mmHg respectively without calibration. Equivalent MAEs on external test datasets without calibration ranged from 15.0 to 25.1 mmHg (SBP) and 7.0 to 10.4 mmHg (DBP). Our results indicate that the performance is strongly influenced by the differences in BP distributions between datasets. We investigated a simple way of improving performance through sample-based domain adaptation and put forward recommendations for training models with good generalization properties. With this work, we hope to educate more researchers for the importance and challenges of out-of-distribution generalization.
Benchmarking machine learning for bowel sound pattern classification from tabular features to pretrained models
Feb 21, 2025Abstract:The development of electronic stethoscopes and wearable recording sensors opened the door to the automated analysis of bowel sound (BS) signals. This enables a data-driven analysis of bowel sound patterns, their interrelations, and their correlation to different pathologies. This work leverages a BS dataset collected from 16 healthy subjects that was annotated according to four established BS patterns. This dataset is used to evaluate the performance of machine learning models to detect and/or classify BS patterns. The selection of considered models covers models using tabular features, convolutional neural networks based on spectrograms and models pre-trained on large audio datasets. The results highlight the clear superiority of pre-trained models, particularly in detecting classes with few samples, achieving an AUC of 0.89 in distinguishing BS from non-BS using a HuBERT model and an AUC of 0.89 in differentiating bowel sound patterns using a Wav2Vec 2.0 model. These results pave the way for an improved understanding of bowel sounds in general and future machine-learning-driven diagnostic applications for gastrointestinal examinations
Explainable and externally validated machine learning for neuropsychiatric diagnosis via electrocardiograms
Feb 07, 2025



Abstract:Electrocardiogram (ECG) analysis has emerged as a promising tool for identifying physiological changes associated with neuropsychiatric conditions. The relationship between cardiovascular health and neuropsychiatric disorders suggests that ECG abnormalities could serve as valuable biomarkers for more efficient detection, therapy monitoring, and risk stratification. However, the potential of the ECG to accurately distinguish neuropsychiatric conditions, particularly among diverse patient populations, remains underexplored. This study utilized ECG markers and basic demographic data to predict neuropsychiatric conditions using machine learning models, with targets defined through ICD-10 codes. Both internal and external validation were performed using the MIMIC-IV and ECG-View datasets respectively. Performance was assessed using AUROC scores. To enhance model interpretability, Shapley values were applied to provide insights into the contributions of individual ECG features to the predictions. Significant predictive performance was observed for conditions within the neurological and psychiatric groups. For the neurological group, Alzheimer's disease (G30) achieved an internal AUROC of 0.813 (0.812-0.814) and an external AUROC of 0.868 (0.867-0.868). In the psychiatric group, unspecified dementia (F03) showed an internal AUROC of 0.849 (0.848-0.849) and an external AUROC of 0.862 (0.861-0.863). Discriminative features align with known ECG markers but also provide hints on potentially new markers. ECG offers significant promise for diagnosing and monitoring neuropsychiatric conditions, with robust predictive performance across internal and external cohorts. Future work should focus on addressing potential confounders, such as therapy-related cardiotoxicity, and expanding the scope of ECG applications, including personalized care and early intervention strategies.
Explainable machine learning for neoplasms diagnosis via electrocardiograms: an externally validated study
Dec 10, 2024



Abstract:Background: Neoplasms remains a leading cause of mortality worldwide, with timely diagnosis being crucial for improving patient outcomes. Current diagnostic methods are often invasive, costly, and inaccessible to many populations. Electrocardiogram (ECG) data, widely available and non-invasive, has the potential to serve as a tool for neoplasms diagnosis by using physiological changes in cardiovascular function associated with neoplastic prescences. Methods: This study explores the application of machine learning models to analyze ECG features for the diagnosis of neoplasms. We developed a pipeline integrating tree-based models with Shapley values for explainability. The model was trained and internally validated and externally validated on a second large-scale independent external cohort to ensure robustness and generalizability. Findings: The results demonstrate that ECG data can effectively capture neoplasms-associated cardiovascular changes, achieving high performance in both internal testing and external validation cohorts. Shapley values identified key ECG features influencing model predictions, revealing established and novel cardiovascular markers linked to neoplastic conditions. This non-invasive approach provides a cost-effective and scalable alternative for the diagnosis of neoplasms, particularly in resource-limited settings. Similarly, useful for the management of secondary cardiovascular effects given neoplasms therapies. Interpretation: This study highlights the feasibility of leveraging ECG signals and machine learning to enhance neoplasms diagnostics. By offering interpretable insights into cardio-neoplasms interactions, this approach bridges existing gaps in non-invasive diagnostics and has implications for integrating ECG-based tools into broader neoplasms diagnostic frameworks, as well as neoplasms therapy management.
Beyond Scalars: Concept-Based Alignment Analysis in Vision Transformers
Dec 09, 2024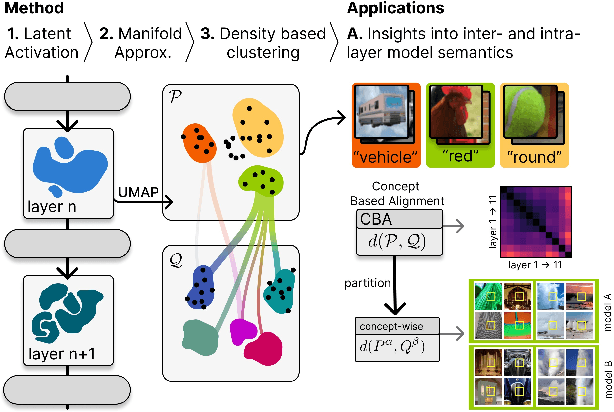
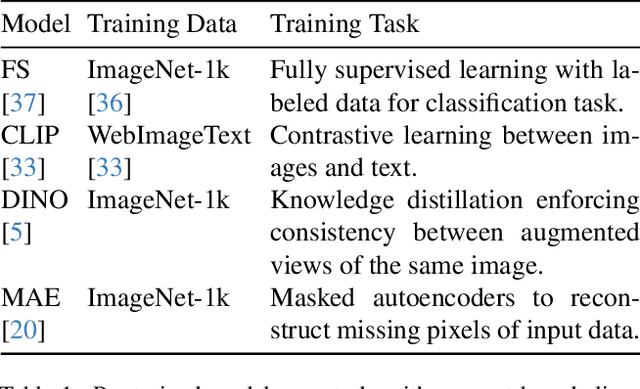
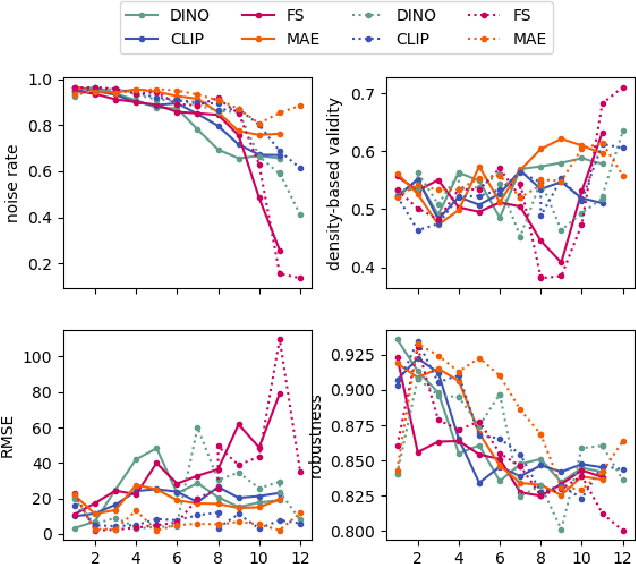
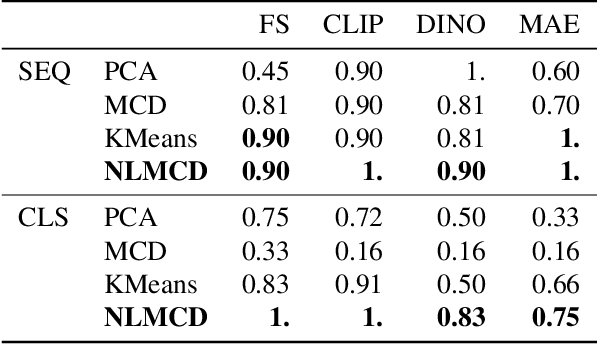
Abstract:Vision transformers (ViTs) can be trained using various learning paradigms, from fully supervised to self-supervised. Diverse training protocols often result in significantly different feature spaces, which are usually compared through alignment analysis. However, current alignment measures quantify this relationship in terms of a single scalar value, obscuring the distinctions between common and unique features in pairs of representations that share the same scalar alignment. We address this limitation by combining alignment analysis with concept discovery, which enables a breakdown of alignment into single concepts encoded in feature space. This fine-grained comparison reveals both universal and unique concepts across different representations, as well as the internal structure of concepts within each of them. Our methodological contributions address two key prerequisites for concept-based alignment: 1) For a description of the representation in terms of concepts that faithfully capture the geometry of the feature space, we define concepts as the most general structure they can possibly form - arbitrary manifolds, allowing hidden features to be described by their proximity to these manifolds. 2) To measure distances between concept proximity scores of two representations, we use a generalized Rand index and partition it for alignment between pairs of concepts. We confirm the superiority of our novel concept definition for alignment analysis over existing linear baselines in a sanity check. The concept-based alignment analysis of representations from four different ViTs reveals that increased supervision correlates with a reduction in the semantic structure of learned representations.
Electrocardiogram-based diagnosis of liver diseases: an externally validated and explainable machine learning approach
Dec 04, 2024



Abstract:Background: Liver diseases are a major global health concern, often diagnosed using resource-intensive methods. Electrocardiogram (ECG) data, widely accessible and non-invasive, offers potential as a diagnostic tool for liver diseases, leveraging the physiological connections between cardiovascular and hepatic health. Methods: This study applies machine learning models to ECG data for the diagnosis of liver diseases. The pipeline, combining tree-based models with Shapley values for explainability, was trained, internally validated, and externally validated on an independent cohort, demonstrating robust generalizability. Findings: Our results demonstrate the potential of ECG to derive biomarkers to diagnose liver diseases. Shapley values revealed key ECG features contributing to model predictions, highlighting already known connections between cardiovascular biomarkers and hepatic conditions as well as providing new ones. Furthermore, our approach holds promise as a scalable and affordable solution for liver disease detection, particularly in resource-limited settings. Interpretation: This study underscores the feasibility of leveraging ECG features and machine learning to enhance the diagnosis of liver diseases. By providing interpretable insights into cardiovascular-liver interactions, the approach bridges existing gaps in non-invasive diagnostics, offering implications for broader systemic disease monitoring.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge