Wilhelm Haverkamp
Explainable and externally validated machine learning for neuropsychiatric diagnosis via electrocardiograms
Feb 07, 2025



Abstract:Electrocardiogram (ECG) analysis has emerged as a promising tool for identifying physiological changes associated with neuropsychiatric conditions. The relationship between cardiovascular health and neuropsychiatric disorders suggests that ECG abnormalities could serve as valuable biomarkers for more efficient detection, therapy monitoring, and risk stratification. However, the potential of the ECG to accurately distinguish neuropsychiatric conditions, particularly among diverse patient populations, remains underexplored. This study utilized ECG markers and basic demographic data to predict neuropsychiatric conditions using machine learning models, with targets defined through ICD-10 codes. Both internal and external validation were performed using the MIMIC-IV and ECG-View datasets respectively. Performance was assessed using AUROC scores. To enhance model interpretability, Shapley values were applied to provide insights into the contributions of individual ECG features to the predictions. Significant predictive performance was observed for conditions within the neurological and psychiatric groups. For the neurological group, Alzheimer's disease (G30) achieved an internal AUROC of 0.813 (0.812-0.814) and an external AUROC of 0.868 (0.867-0.868). In the psychiatric group, unspecified dementia (F03) showed an internal AUROC of 0.849 (0.848-0.849) and an external AUROC of 0.862 (0.861-0.863). Discriminative features align with known ECG markers but also provide hints on potentially new markers. ECG offers significant promise for diagnosing and monitoring neuropsychiatric conditions, with robust predictive performance across internal and external cohorts. Future work should focus on addressing potential confounders, such as therapy-related cardiotoxicity, and expanding the scope of ECG applications, including personalized care and early intervention strategies.
Explainable machine learning for neoplasms diagnosis via electrocardiograms: an externally validated study
Dec 10, 2024



Abstract:Background: Neoplasms remains a leading cause of mortality worldwide, with timely diagnosis being crucial for improving patient outcomes. Current diagnostic methods are often invasive, costly, and inaccessible to many populations. Electrocardiogram (ECG) data, widely available and non-invasive, has the potential to serve as a tool for neoplasms diagnosis by using physiological changes in cardiovascular function associated with neoplastic prescences. Methods: This study explores the application of machine learning models to analyze ECG features for the diagnosis of neoplasms. We developed a pipeline integrating tree-based models with Shapley values for explainability. The model was trained and internally validated and externally validated on a second large-scale independent external cohort to ensure robustness and generalizability. Findings: The results demonstrate that ECG data can effectively capture neoplasms-associated cardiovascular changes, achieving high performance in both internal testing and external validation cohorts. Shapley values identified key ECG features influencing model predictions, revealing established and novel cardiovascular markers linked to neoplastic conditions. This non-invasive approach provides a cost-effective and scalable alternative for the diagnosis of neoplasms, particularly in resource-limited settings. Similarly, useful for the management of secondary cardiovascular effects given neoplasms therapies. Interpretation: This study highlights the feasibility of leveraging ECG signals and machine learning to enhance neoplasms diagnostics. By offering interpretable insights into cardio-neoplasms interactions, this approach bridges existing gaps in non-invasive diagnostics and has implications for integrating ECG-based tools into broader neoplasms diagnostic frameworks, as well as neoplasms therapy management.
Electrocardiogram-based diagnosis of liver diseases: an externally validated and explainable machine learning approach
Dec 04, 2024



Abstract:Background: Liver diseases are a major global health concern, often diagnosed using resource-intensive methods. Electrocardiogram (ECG) data, widely accessible and non-invasive, offers potential as a diagnostic tool for liver diseases, leveraging the physiological connections between cardiovascular and hepatic health. Methods: This study applies machine learning models to ECG data for the diagnosis of liver diseases. The pipeline, combining tree-based models with Shapley values for explainability, was trained, internally validated, and externally validated on an independent cohort, demonstrating robust generalizability. Findings: Our results demonstrate the potential of ECG to derive biomarkers to diagnose liver diseases. Shapley values revealed key ECG features contributing to model predictions, highlighting already known connections between cardiovascular biomarkers and hepatic conditions as well as providing new ones. Furthermore, our approach holds promise as a scalable and affordable solution for liver disease detection, particularly in resource-limited settings. Interpretation: This study underscores the feasibility of leveraging ECG features and machine learning to enhance the diagnosis of liver diseases. By providing interpretable insights into cardiovascular-liver interactions, the approach bridges existing gaps in non-invasive diagnostics, offering implications for broader systemic disease monitoring.
Cardiac and extracardiac discharge diagnosis prediction from emergency department ECGs using deep learning
Dec 18, 2023



Abstract:Current deep learning algorithms designed for automatic ECG analysis have exhibited notable accuracy. However, akin to traditional electrocardiography, they tend to be narrowly focused and typically address a singular diagnostic condition. In this study, we specifically demonstrate the capability of a single model to predict a diverse range of both cardiac and non-cardiac discharge diagnoses based on a sole ECG collected in the emergency department. Among the 1,076 hierarchically structured ICD codes considered, our model achieves an AUROC exceeding 0.8 in 439 of them. This underscores the models proficiency in handling a wide array of diagnostic scenarios. We emphasize the potential of utilizing this model as a screening tool, potentially integrated into a holistic clinical decision support system for efficiently triaging patients in the emergency department. This research underscores the remarkable capabilities of comprehensive ECG analysis algorithms and the extensive range of possibilities facilitated by the open MIMIC-IV-ECG dataset. Finally, our data may play a pivotal role in revolutionizing the way ECG analysis is performed, marking a significant advancement in the field.
Uncovering ECG Changes during Healthy Aging using Explainable AI
Oct 11, 2023



Abstract:Cardiovascular diseases remain the leading global cause of mortality. This necessitates a profound understanding of heart aging processes to diagnose constraints in cardiovascular fitness. Traditionally, most of such insights have been drawn from the analysis of electrocardiogram (ECG) feature changes of individuals as they age. However, these features, while informative, may potentially obscure underlying data relationships. In this paper, we employ a deep-learning model and a tree-based model to analyze ECG data from a robust dataset of healthy individuals across varying ages in both raw signals and ECG feature format. Explainable AI techniques are then used to identify ECG features or raw signal characteristics are most discriminative for distinguishing between age groups. Our analysis with tree-based classifiers reveal age-related declines in inferred breathing rates and identifies notably high SDANN values as indicative of elderly individuals, distinguishing them from younger adults. Furthermore, the deep-learning model underscores the pivotal role of the P-wave in age predictions across all age groups, suggesting potential changes in the distribution of different P-wave types with age. These findings shed new light on age-related ECG changes, offering insights that transcend traditional feature-based approaches.
Explaining Deep Learning for ECG Analysis: Building Blocks for Auditing and Knowledge Discovery
May 26, 2023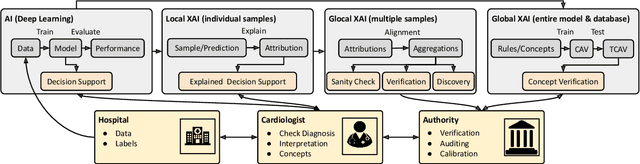
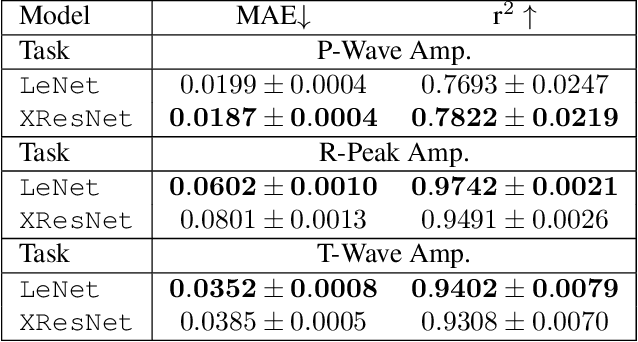
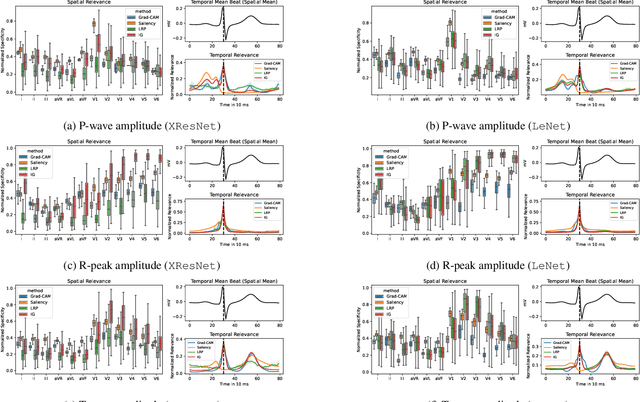
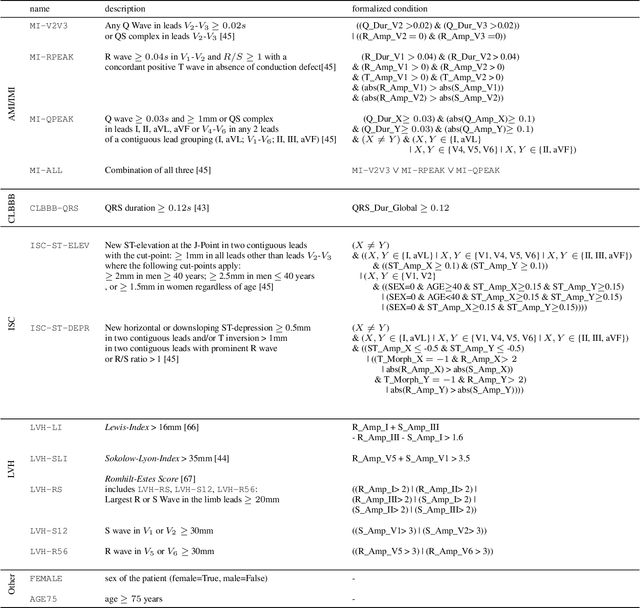
Abstract:Deep neural networks have become increasingly popular for analyzing ECG data because of their ability to accurately identify cardiac conditions and hidden clinical factors. However, the lack of transparency due to the black box nature of these models is a common concern. To address this issue, explainable AI (XAI) methods can be employed. In this study, we present a comprehensive analysis of post-hoc XAI methods, investigating the local (attributions per sample) and global (based on domain expert concepts) perspectives. We have established a set of sanity checks to identify sensible attribution methods, and we provide quantitative evidence in accordance with expert rules. This dataset-wide analysis goes beyond anecdotal evidence by aggregating data across patient subgroups. Furthermore, we demonstrate how these XAI techniques can be utilized for knowledge discovery, such as identifying subtypes of myocardial infarction. We believe that these proposed methods can serve as building blocks for a complementary assessment of the internal validity during a certification process, as well as for knowledge discovery in the field of ECG analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge