Lihua Li
DML-GANR: Deep Metric Learning With Generative Adversarial Network Regularization for High Spatial Resolution Remote Sensing Image Retrieval
Oct 07, 2020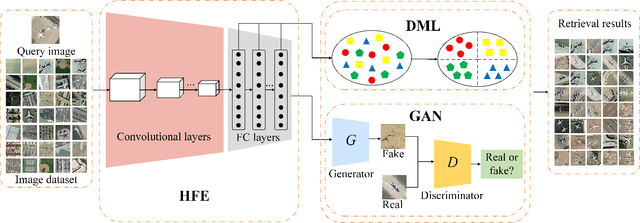
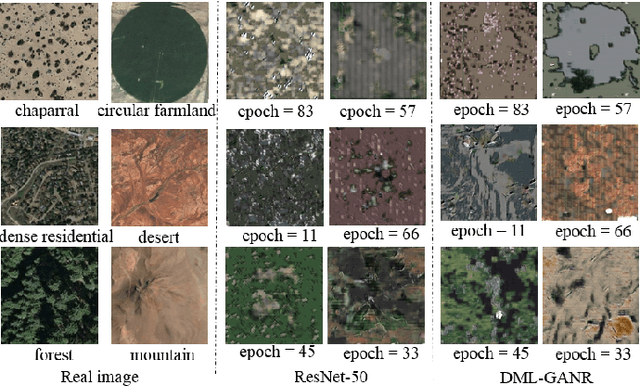
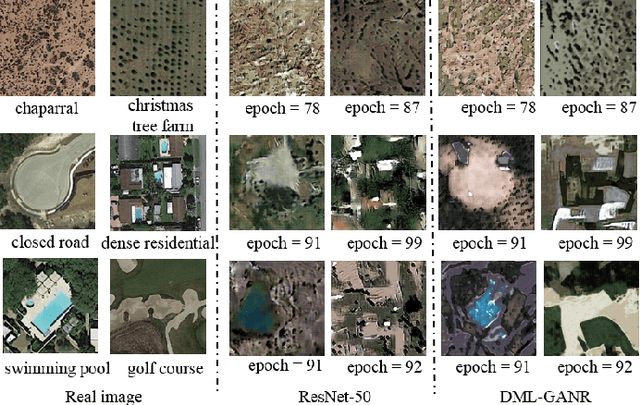
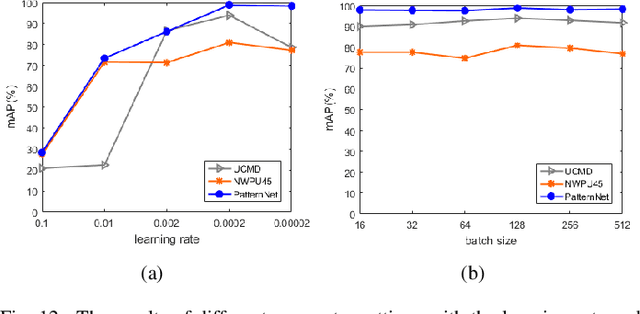
Abstract:With a small number of labeled samples for training, it can save considerable manpower and material resources, especially when the amount of high spatial resolution remote sensing images (HSR-RSIs) increases considerably. However, many deep models face the problem of overfitting when using a small number of labeled samples. This might degrade HSRRSI retrieval accuracy. Aiming at obtaining more accurate HSR-RSI retrieval performance with small training samples, we develop a deep metric learning approach with generative adversarial network regularization (DML-GANR) for HSR-RSI retrieval. The DML-GANR starts from a high-level feature extraction (HFE) to extract high-level features, which includes convolutional layers and fully connected (FC) layers. Each of the FC layers is constructed by deep metric learning (DML) to maximize the interclass variations and minimize the intraclass variations. The generative adversarial network (GAN) is adopted to mitigate the overfitting problem and validate the qualities of extracted high-level features. DML-GANR is optimized through a customized approach, and the optimal parameters are obtained. The experimental results on the three data sets demonstrate the superior performance of DML-GANR over state-of-the-art techniques in HSR-RSI retrieval.
SLCRF: Subspace Learning with Conditional Random Field for Hyperspectral Image Classification
Oct 07, 2020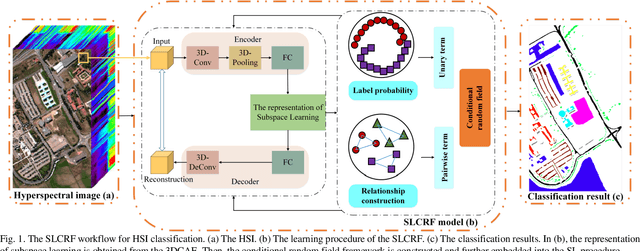

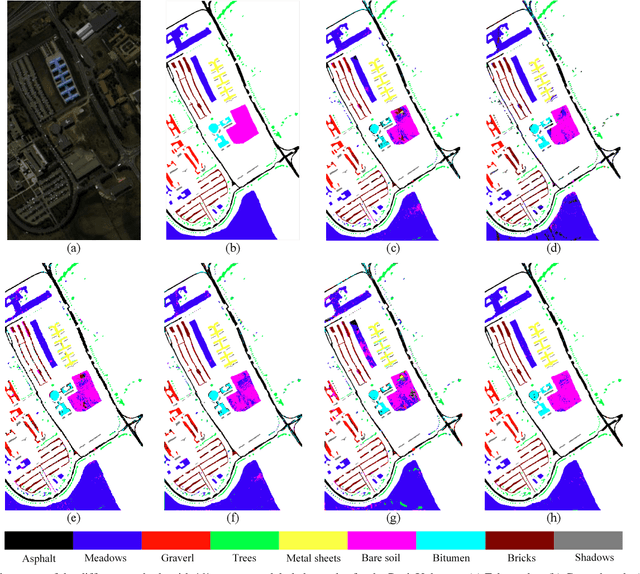
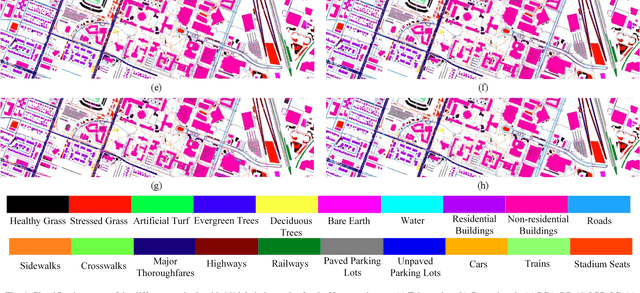
Abstract:Subspace learning (SL) plays an important role in hyperspectral image (HSI) classification, since it can provide an effective solution to reduce the redundant information in the image pixels of HSIs. Previous works about SL aim to improve the accuracy of HSI recognition. Using a large number of labeled samples, related methods can train the parameters of the proposed solutions to obtain better representations of HSI pixels. However, the data instances may not be sufficient enough to learn a precise model for HSI classification in real applications. Moreover, it is well-known that it takes much time, labor and human expertise to label HSI images. To avoid the aforementioned problems, a novel SL method that includes the probability assumption called subspace learning with conditional random field (SLCRF) is developed. In SLCRF, first, the 3D convolutional autoencoder (3DCAE) is introduced to remove the redundant information in HSI pixels. In addition, the relationships are also constructed using the spectral-spatial information among the adjacent pixels. Then, the conditional random field (CRF) framework can be constructed and further embedded into the HSI SL procedure with the semi-supervised approach. Through the linearized alternating direction method termed LADMAP, the objective function of SLCRF is optimized using a defined iterative algorithm. The proposed method is comprehensively evaluated using the challenging public HSI datasets. We can achieve stateof-the-art performance using these HSI sets.
DLBI: Deep learning guided Bayesian inference for structure reconstruction of super-resolution fluorescence microscopy
Sep 01, 2018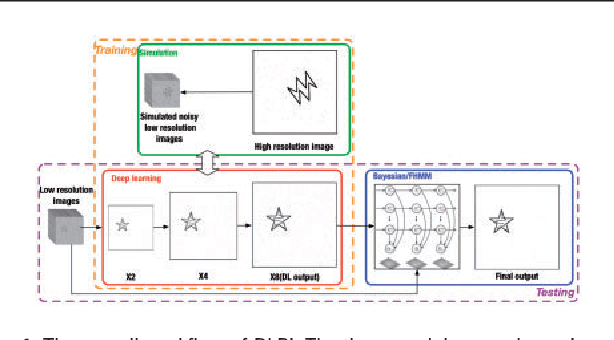
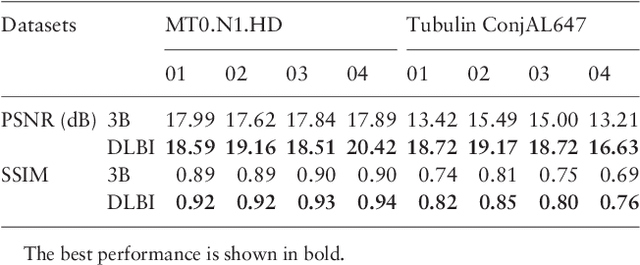
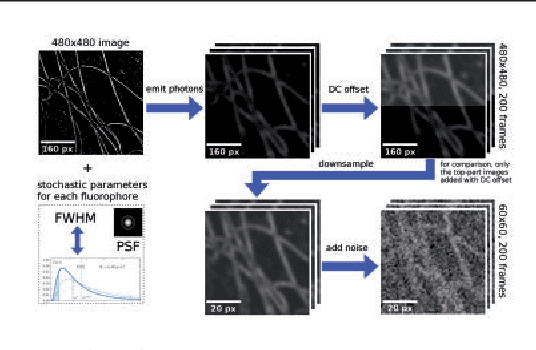
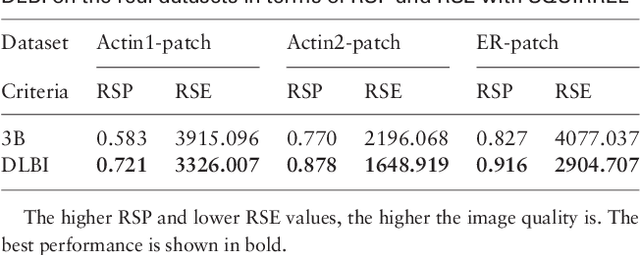
Abstract:Super-resolution fluorescence microscopy, with a resolution beyond the diffraction limit of light, has become an indispensable tool to directly visualize biological structures in living cells at a nanometer-scale resolution. Despite advances in high-density super-resolution fluorescent techniques, existing methods still have bottlenecks, including extremely long execution time, artificial thinning and thickening of structures, and lack of ability to capture latent structures. Here we propose a novel deep learning guided Bayesian inference approach, DLBI, for the time-series analysis of high-density fluorescent images. Our method combines the strength of deep learning and statistical inference, where deep learning captures the underlying distribution of the fluorophores that are consistent with the observed time-series fluorescent images by exploring local features and correlation along time-axis, and statistical inference further refines the ultrastructure extracted by deep learning and endues physical meaning to the final image. Comprehensive experimental results on both real and simulated datasets demonstrate that our method provides more accurate and realistic local patch and large-field reconstruction than the state-of-the-art method, the 3B analysis, while our method is more than two orders of magnitude faster. The main program is available at https://github.com/lykaust15/DLBI
* Accepted by ISMB 2018
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge