Kevin He
AI as a Teaching Partner: Early Lessons from Classroom Codesign with Secondary Teachers
Dec 12, 2025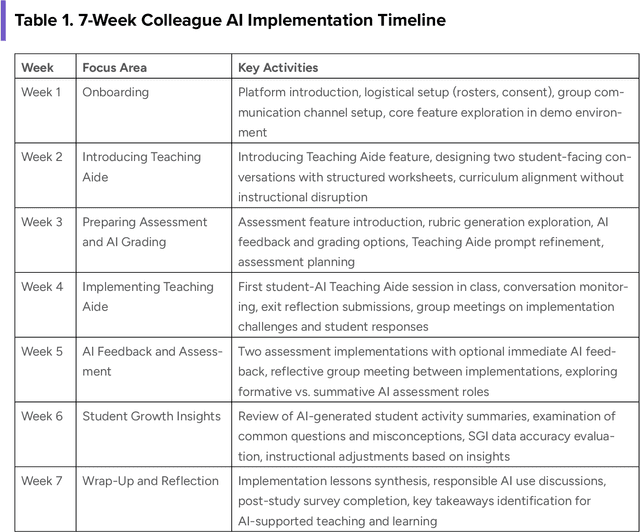
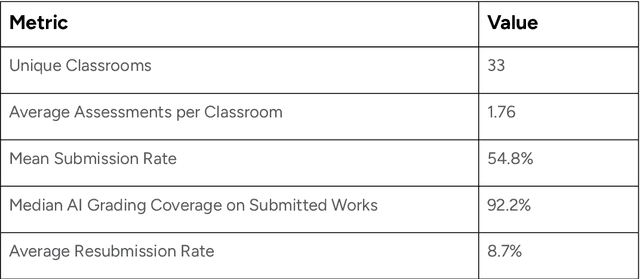
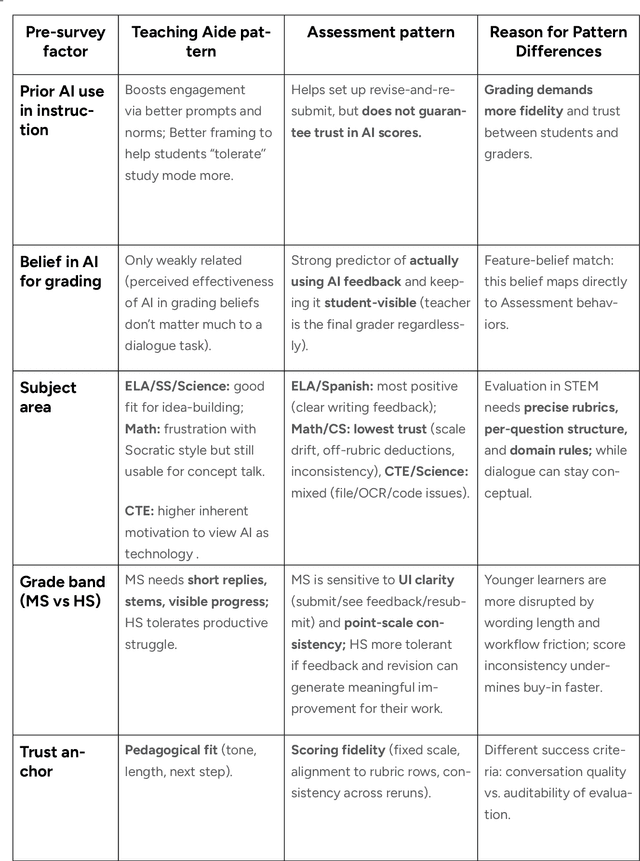
Abstract:This report presents a comprehensive account of the Colleague AI Classroom pilot, a collaborative design (co-design) study that brought generative AI technology directly into real classrooms. In this study, AI functioned as a third agent, an active participant that mediated feedback, supported inquiry, and extended teachers' instructional reach while preserving human judgment and teacher authority. Over seven weeks in spring 2025, 21 in-service teachers from four Washington State public school districts and one independent school integrated four AI-powered features of the Colleague AI Classroom into their instruction: Teaching Aide, Assessment and AI Grading, AI Tutor, and Student Growth Insights. More than 600 students in grades 6-12 used the platform in class at the direction of their teachers, who designed and facilitated the AI activities. During the Classroom pilot, teachers were co-design partners: they planned activities, implemented them with students, and provided weekly reflections on AI's role in classroom settings. The teachers' feedback guided iterative improvements for Colleague AI. The research team captured rich data through surveys, planning and reflection forms, group meetings, one-on-one interviews, and platform usage logs to understand where AI adds instructional value and where it requires refinement.
Decoding Instructional Dialogue: Human-AI Collaborative Analysis of Teacher Use of AI Tool at Scale
Jul 23, 2025Abstract:The integration of large language models (LLMs) into educational tools has the potential to substantially impact how teachers plan instruction, support diverse learners, and engage in professional reflection. Yet little is known about how educators actually use these tools in practice and how their interactions with AI can be meaningfully studied at scale. This paper presents a human-AI collaborative methodology for large-scale qualitative analysis of over 140,000 educator-AI messages drawn from a generative AI platform used by K-12 teachers. Through a four-phase coding pipeline, we combined inductive theme discovery, codebook development, structured annotation, and model benchmarking to examine patterns of educator engagement and evaluate the performance of LLMs in qualitative coding tasks. We developed a hierarchical codebook aligned with established teacher evaluation frameworks, capturing educators' instructional goals, contextual needs, and pedagogical strategies. Our findings demonstrate that LLMs, particularly Claude 3.5 Haiku, can reliably support theme identification, extend human recognition in complex scenarios, and outperform open-weight models in both accuracy and structural reliability. The analysis also reveals substantive patterns in how educators inquire AI to enhance instructional practices (79.7 percent of total conversations), create or adapt content (76.1 percent), support assessment and feedback loop (46.9 percent), attend to student needs for tailored instruction (43.3 percent), and assist other professional responsibilities (34.2 percent), highlighting emerging AI-related competencies that have direct implications for teacher preparation and professional development. This study offers a scalable, transparent model for AI-augmented qualitative research and provides foundational insights into the evolving role of generative AI in educational practice.
Human Misperception of Generative-AI Alignment: A Laboratory Experiment
Feb 20, 2025Abstract:We conduct an incentivized laboratory experiment to study people's perception of generative artificial intelligence (GenAI) alignment in the context of economic decision-making. Using a panel of economic problems spanning the domains of risk, time preference, social preference, and strategic interactions, we ask human subjects to make choices for themselves and to predict the choices made by GenAI on behalf of a human user. We find that people overestimate the degree of alignment between GenAI's choices and human choices. In every problem, human subjects' average prediction about GenAI's choice is substantially closer to the average human-subject choice than it is to the GenAI choice. At the individual level, different subjects' predictions about GenAI's choice in a given problem are highly correlated with their own choices in the same problem. We explore the implications of people overestimating GenAI alignment in a simple theoretical model.
High-Throughput SAT Sampling
Feb 12, 2025Abstract:In this work, we present a novel technique for GPU-accelerated Boolean satisfiability (SAT) sampling. Unlike conventional sampling algorithms that directly operate on conjunctive normal form (CNF), our method transforms the logical constraints of SAT problems by factoring their CNF representations into simplified multi-level, multi-output Boolean functions. It then leverages gradient-based optimization to guide the search for a diverse set of valid solutions. Our method operates directly on the circuit structure of refactored SAT instances, reinterpreting the SAT problem as a supervised multi-output regression task. This differentiable technique enables independent bit-wise operations on each tensor element, allowing parallel execution of learning processes. As a result, we achieve GPU-accelerated sampling with significant runtime improvements ranging from $33.6\times$ to $523.6\times$ over state-of-the-art heuristic samplers. We demonstrate the superior performance of our sampling method through an extensive evaluation on $60$ instances from a public domain benchmark suite utilized in previous studies.
Training-Aware Risk Control for Intensity Modulated Radiation Therapies Quality Assurance with Conformal Prediction
Jan 15, 2025Abstract:Measurement quality assurance (QA) practices play a key role in the safe use of Intensity Modulated Radiation Therapies (IMRT) for cancer treatment. These practices have reduced measurement-based IMRT QA failure below 1%. However, these practices are time and labor intensive which can lead to delays in patient care. In this study, we examine how conformal prediction methodologies can be used to robustly triage plans. We propose a new training-aware conformal risk control method by combining the benefit of conformal risk control and conformal training. We incorporate the decision making thresholds based on the gamma passing rate, along with the risk functions used in clinical evaluation, into the design of the risk control framework. Our method achieves high sensitivity and specificity and significantly reduces the number of plans needing measurement without generating a huge confidence interval. Our results demonstrate the validity and applicability of conformal prediction methods for improving efficiency and reducing the workload of the IMRT QA process.
KL-divergence Based Deep Learning for Discrete Time Model
Aug 10, 2022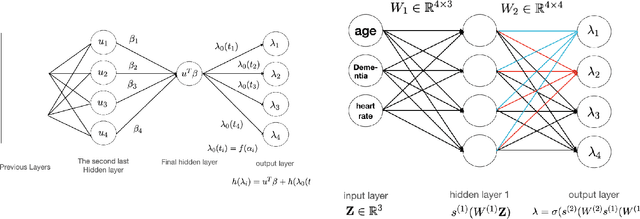
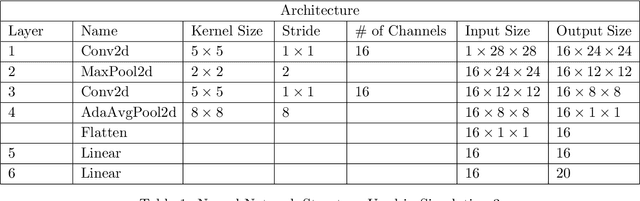
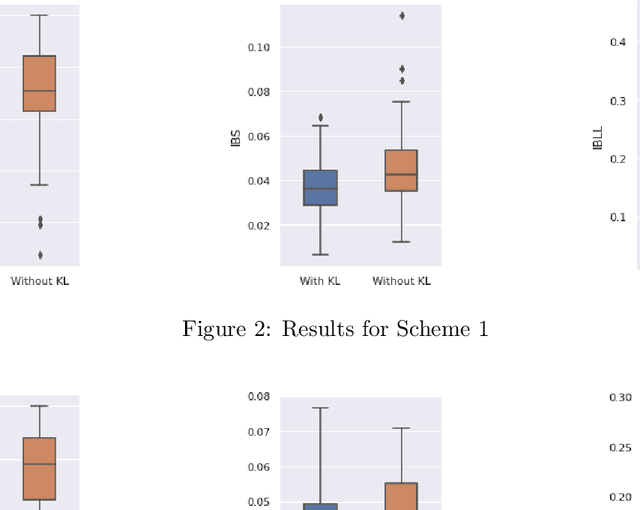
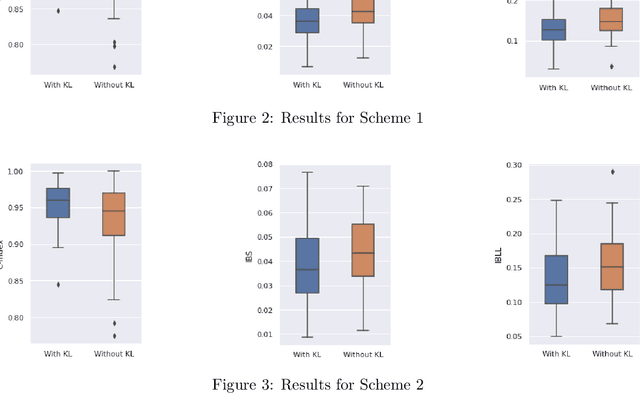
Abstract:Neural Network (Deep Learning) is a modern model in Artificial Intelligence and it has been exploited in Survival Analysis. Although several improvements have been shown by previous works, training an excellent deep learning model requires a huge amount of data, which may not hold in practice. To address this challenge, we develop a Kullback-Leibler-based (KL) deep learning procedure to integrate external survival prediction models with newly collected time-to-event data. Time-dependent KL discrimination information is utilized to measure the discrepancy between the external and internal data. This is the first work considering using prior information to deal with short data problem in Survival Analysis for deep learning. Simulation and real data results show that the proposed model achieves better performance and higher robustness compared with previous works.
Covariance-Insured Screening
May 17, 2018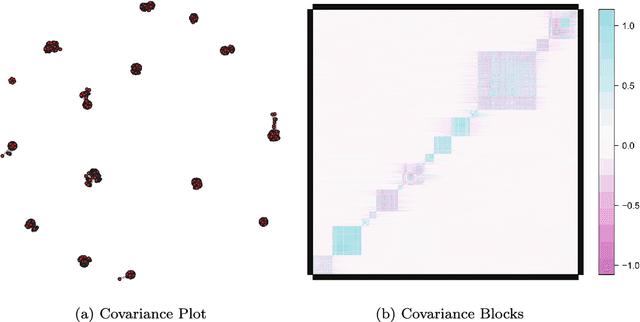
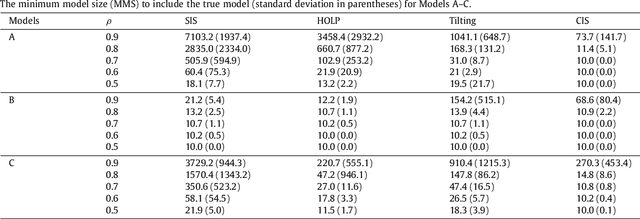
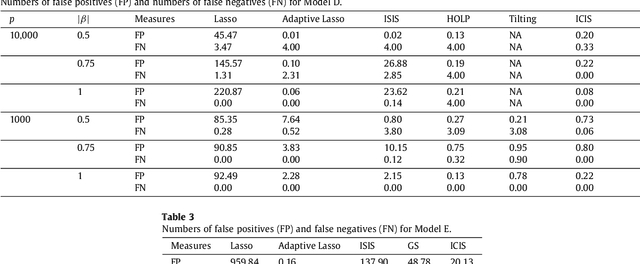
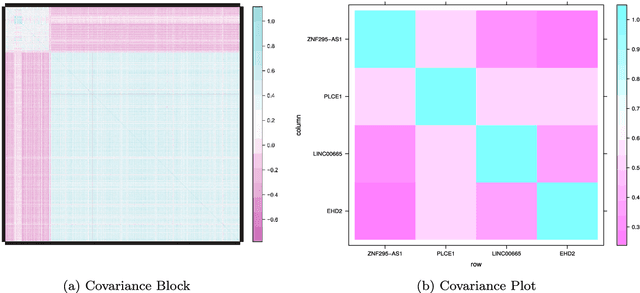
Abstract:Modern bio-technologies have produced a vast amount of high-throughput data with the number of predictors far greater than the sample size. In order to identify more novel biomarkers and understand biological mechanisms, it is vital to detect signals weakly associated with outcomes among ultrahigh-dimensional predictors. However, existing screening methods, which typically ignore correlation information, are likely to miss these weak signals. By incorporating the inter-feature dependence, we propose a covariance-insured screening methodology to identify predictors that are jointly informative but only marginally weakly associated with outcomes. The validity of the method is examined via extensive simulations and real data studies for selecting potential genetic factors related to the onset of cancer.
Classification with Ultrahigh-Dimensional Features
Nov 04, 2016



Abstract:Although much progress has been made in classification with high-dimensional features \citep{Fan_Fan:2008, JGuo:2010, CaiSun:2014, PRXu:2014}, classification with ultrahigh-dimensional features, wherein the features much outnumber the sample size, defies most existing work. This paper introduces a novel and computationally feasible multivariate screening and classification method for ultrahigh-dimensional data. Leveraging inter-feature correlations, the proposed method enables detection of marginally weak and sparse signals and recovery of the true informative feature set, and achieves asymptotic optimal misclassification rates. We also show that the proposed procedure provides more powerful discovery boundaries compared to those in \citet{CaiSun:2014} and \citet{JJin:2009}. The performance of the proposed procedure is evaluated using simulation studies and demonstrated via classification of patients with different post-transplantation renal functional types.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge