Kaiwei Sun
AEMLO: AutoEncoder-Guided Multi-Label Oversampling
Aug 23, 2024Abstract:Class imbalance significantly impacts the performance of multi-label classifiers. Oversampling is one of the most popular approaches, as it augments instances associated with less frequent labels to balance the class distribution. Existing oversampling methods generate feature vectors of synthetic samples through replication or linear interpolation and assign labels through neighborhood information. Linear interpolation typically generates new samples between existing data points, which may result in insufficient diversity of synthesized samples and further lead to the overfitting issue. Deep learning-based methods, such as AutoEncoders, have been proposed to generate more diverse and complex synthetic samples, achieving excellent performance on imbalanced binary or multi-class datasets. In this study, we introduce AEMLO, an AutoEncoder-guided Oversampling technique specifically designed for tackling imbalanced multi-label data. AEMLO is built upon two fundamental components. The first is an encoder-decoder architecture that enables the model to encode input data into a low-dimensional feature space, learn its latent representations, and then reconstruct it back to its original dimension, thus applying to the generation of new data. The second is an objective function tailored to optimize the sampling task for multi-label scenarios. We show that AEMLO outperforms the existing state-of-the-art methods with extensive empirical studies.
Fine-Grained Selective Similarity Integration for Drug-Target Interaction Prediction
Dec 01, 2022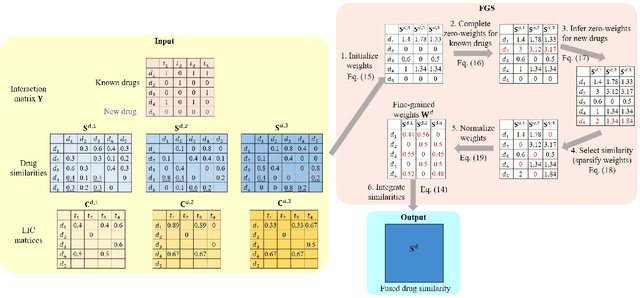
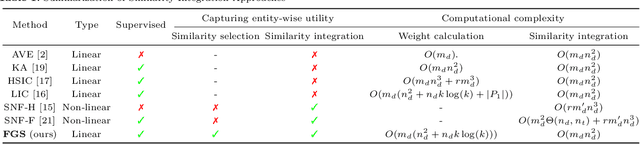
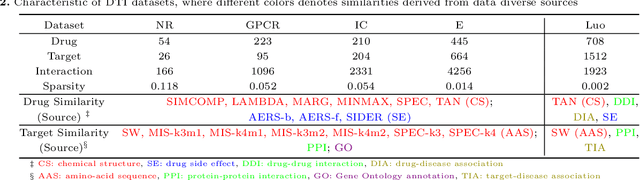
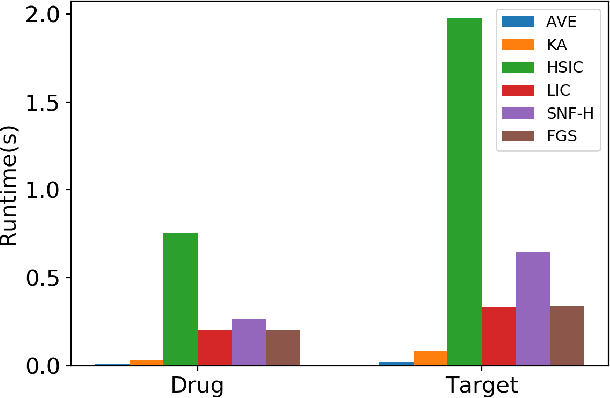
Abstract:The discovery of drug-target interactions (DTIs) is a pivotal process in pharmaceutical development. Computational approaches are a promising and efficient alternative to tedious and costly wet-lab experiments for predicting novel DTIs from numerous candidates. Recently, with the availability of abundant heterogeneous biological information from diverse data sources, computational methods have been able to leverage multiple drug and target similarities to boost the performance of DTI prediction. Similarity integration is an effective and flexible strategy to extract crucial information across complementary similarity views, providing a compressed input for any similarity-based DTI prediction model. However, existing similarity integration methods filter and fuse similarities from a global perspective, neglecting the utility of similarity views for each drug and target. In this study, we propose a Fine-Grained Selective similarity integration approach, called FGS, which employs a local interaction consistency-based weight matrix to capture and exploit the importance of similarities at a finer granularity in both similarity selection and combination steps. We evaluate FGS on five DTI prediction datasets under various prediction settings. Experimental results show that our method not only outperforms similarity integration competitors with comparable computational costs, but also achieves better prediction performance than state-of-the-art DTI prediction approaches by collaborating with conventional base models. Furthermore, case studies on the analysis of similarity weights and on the verification of novel predictions confirm the practical ability of FGS.
AI-enabled Automatic Multimodal Fusion of Cone-Beam CT and Intraoral Scans for Intelligent 3D Tooth-Bone Reconstruction and Clinical Applications
Mar 11, 2022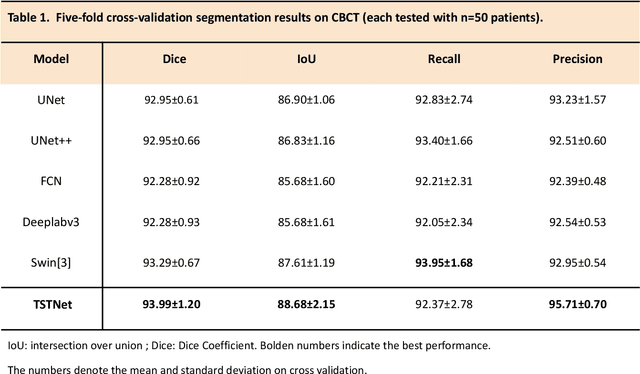
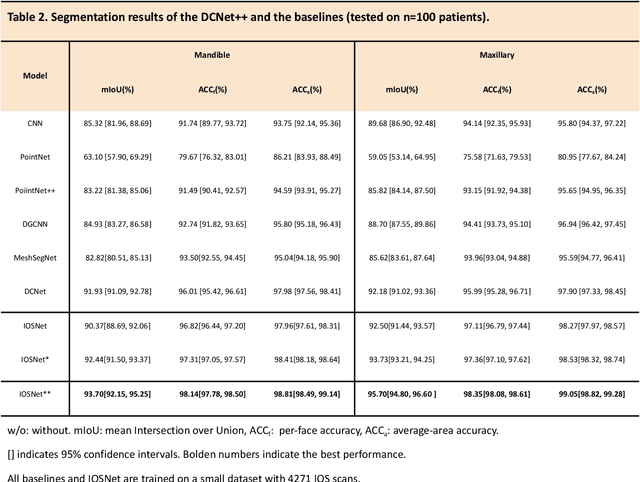

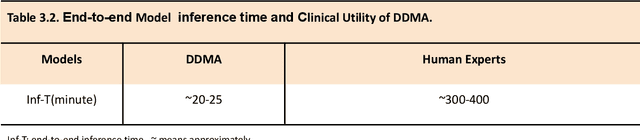
Abstract:A critical step in virtual dental treatment planning is to accurately delineate all tooth-bone structures from CBCT with high fidelity and accurate anatomical information. Previous studies have established several methods for CBCT segmentation using deep learning. However, the inherent resolution discrepancy of CBCT and the loss of occlusal and dentition information largely limited its clinical applicability. Here, we present a Deep Dental Multimodal Analysis (DDMA) framework consisting of a CBCT segmentation model, an intraoral scan (IOS) segmentation model (the most accurate digital dental model), and a fusion model to generate 3D fused crown-root-bone structures with high fidelity and accurate occlusal and dentition information. Our model was trained with a large-scale dataset with 503 CBCT and 28,559 IOS meshes manually annotated by experienced human experts. For CBCT segmentation, we use a five-fold cross validation test, each with 50 CBCT, and our model achieves an average Dice coefficient and IoU of 93.99% and 88.68%, respectively, significantly outperforming the baselines. For IOS segmentations, our model achieves an mIoU of 93.07% and 95.70% on the maxillary and mandible on a test set of 200 IOS meshes, which are 1.77% and 3.52% higher than the state-of-art method. Our DDMA framework takes about 20 to 25 minutes to generate the fused 3D mesh model following the sequential processing order, compared to over 5 hours by human experts. Notably, our framework has been incorporated into a software by a clear aligner manufacturer, and real-world clinical cases demonstrate that our model can visualize crown-root-bone structures during the entire orthodontic treatment and can predict risks like dehiscence and fenestration. These findings demonstrate the potential of multi-modal deep learning to improve the quality of digital dental models and help dentists make better clinical decisions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge