Kai Standvoss
Atlas: A Novel Pathology Foundation Model by Mayo Clinic, Charité, and Aignostics
Jan 10, 2025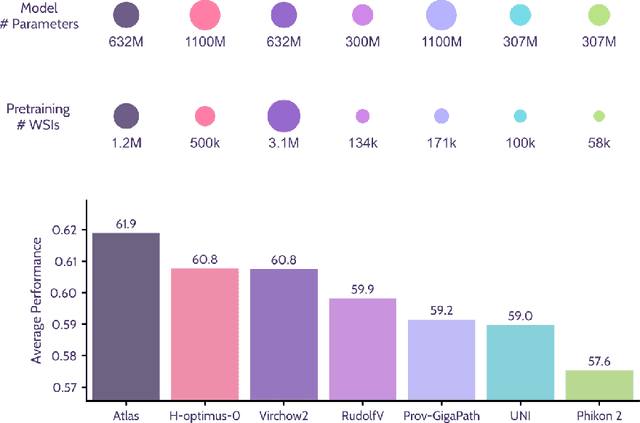
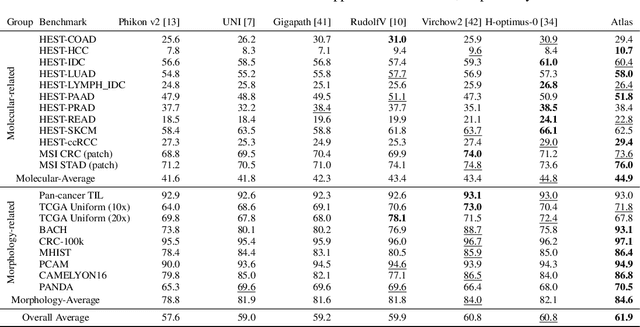
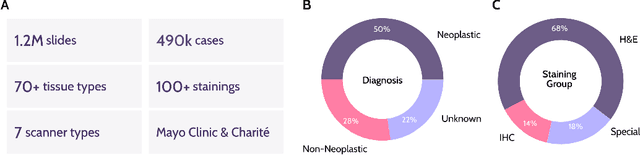
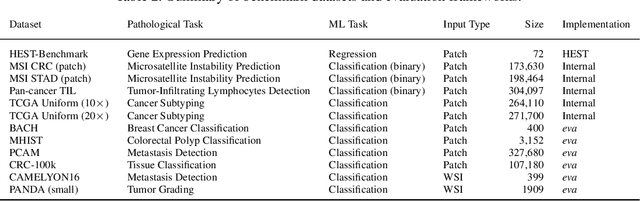
Abstract:Recent advances in digital pathology have demonstrated the effectiveness of foundation models across diverse applications. In this report, we present Atlas, a novel vision foundation model based on the RudolfV approach. Our model was trained on a dataset comprising 1.2 million histopathology whole slide images, collected from two medical institutions: Mayo Clinic and Charit\'e - Universt\"atsmedizin Berlin. Comprehensive evaluations show that Atlas achieves state-of-the-art performance across twenty-one public benchmark datasets, even though it is neither the largest model by parameter count nor by training dataset size.
xCG: Explainable Cell Graphs for Survival Prediction in Non-Small Cell Lung Cancer
Nov 12, 2024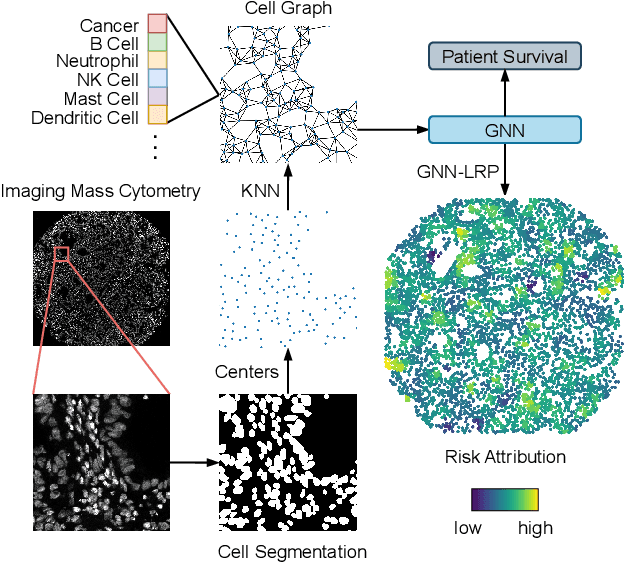
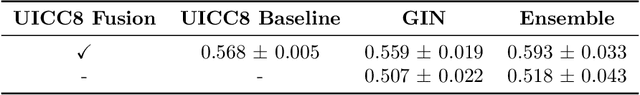
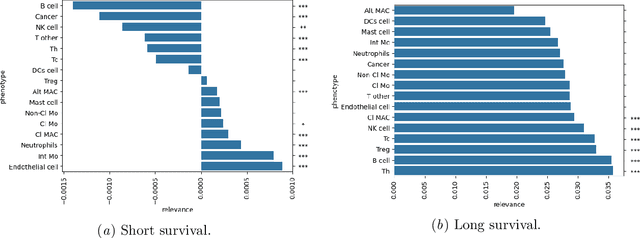
Abstract:Understanding how deep learning models predict oncology patient risk can provide critical insights into disease progression, support clinical decision-making, and pave the way for trustworthy and data-driven precision medicine. Building on recent advances in the spatial modeling of the tumor microenvironment using graph neural networks, we present an explainable cell graph (xCG) approach for survival prediction. We validate our model on a public cohort of imaging mass cytometry (IMC) data for 416 cases of lung adenocarcinoma. We explain survival predictions in terms of known phenotypes on the cell level by computing risk attributions over cell graphs, for which we propose an efficient grid-based layer-wise relevance propagation (LRP) method. Our ablation studies highlight the importance of incorporating the cancer stage and model ensembling to improve the quality of risk estimates. Our xCG method, together with the IMC data, is made publicly available to support further research.
DiffInfinite: Large Mask-Image Synthesis via Parallel Random Patch Diffusion in Histopathology
Jun 23, 2023



Abstract:We present DiffInfinite, a hierarchical diffusion model that generates arbitrarily large histological images while preserving long-range correlation structural information. Our approach first generates synthetic segmentation masks, subsequently used as conditions for the high-fidelity generative diffusion process. The proposed sampling method can be scaled up to any desired image size while only requiring small patches for fast training. Moreover, it can be parallelized more efficiently than previous large-content generation methods while avoiding tiling artefacts. The training leverages classifier-free guidance to augment a small, sparsely annotated dataset with unlabelled data. Our method alleviates unique challenges in histopathological imaging practice: large-scale information, costly manual annotation, and protective data handling. The biological plausibility of DiffInfinite data is validated in a survey by ten experienced pathologists as well as a downstream segmentation task. Furthermore, the model scores strongly on anti-copying metrics which is beneficial for the protection of patient data.
Discourse Embellishment Using a Deep Encoder-Decoder Network
Oct 18, 2018Abstract:We suggest a new NLG task in the context of the discourse generation pipeline of computational storytelling systems. This task, textual embellishment, is defined by taking a text as input and generating a semantically equivalent output with increased lexical and syntactic complexity. Ideally, this would allow the authors of computational storytellers to implement just lightweight NLG systems and use a domain-independent embellishment module to translate its output into more literary text. We present promising first results on this task using LSTM Encoder-Decoder networks trained on the WikiLarge dataset. Furthermore, we introduce "Compiled Computer Tales", a corpus of computationally generated stories, that can be used to test the capabilities of embellishment algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge