Gabriel Dernbach
MapPFN: Learning Causal Perturbation Maps in Context
Jan 28, 2026Abstract:Planning effective interventions in biological systems requires treatment-effect models that adapt to unseen biological contexts by identifying their specific underlying mechanisms. Yet single-cell perturbation datasets span only a handful of biological contexts, and existing methods cannot leverage new interventional evidence at inference time to adapt beyond their training data. To meta-learn a perturbation effect estimator, we present MapPFN, a prior-data fitted network (PFN) pretrained on synthetic data generated from a prior over causal perturbations. Given a set of experiments, MapPFN uses in-context learning to predict post-perturbation distributions, without gradient-based optimization. Despite being pretrained on in silico gene knockouts alone, MapPFN identifies differentially expressed genes, matching the performance of models trained on real single-cell data. Our code and data are available at https://github.com/marvinsxtr/MapPFN.
Atlas 2 - Foundation models for clinical deployment
Jan 08, 2026Abstract:Pathology foundation models substantially advanced the possibilities in computational pathology -- yet tradeoffs in terms of performance, robustness, and computational requirements remained, which limited their clinical deployment. In this report, we present Atlas 2, Atlas 2-B, and Atlas 2-S, three pathology vision foundation models which bridge these shortcomings by showing state-of-the-art performance in prediction performance, robustness, and resource efficiency in a comprehensive evaluation across eighty public benchmarks. Our models were trained on the largest pathology foundation model dataset to date comprising 5.5 million histopathology whole slide images, collected from three medical institutions Charité - Universtätsmedizin Berlin, LMU Munich, and Mayo Clinic.
Atlas: A Novel Pathology Foundation Model by Mayo Clinic, Charité, and Aignostics
Jan 10, 2025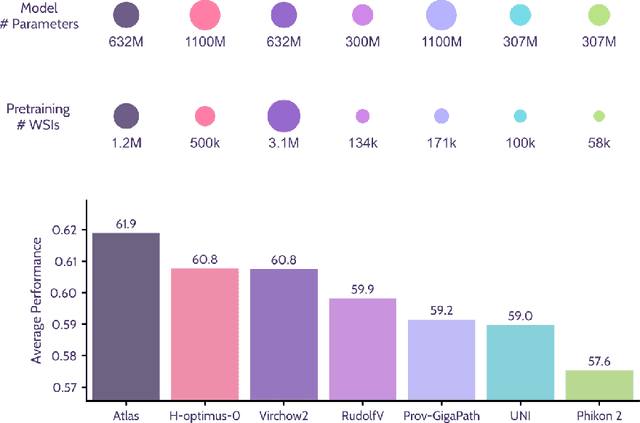
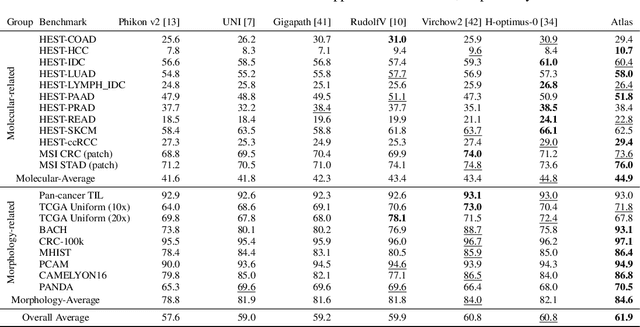
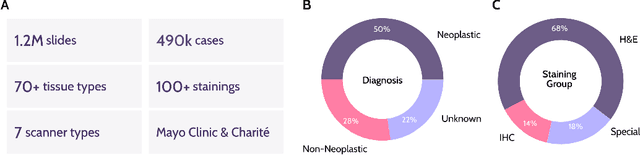
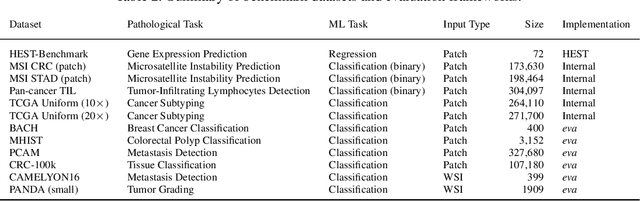
Abstract:Recent advances in digital pathology have demonstrated the effectiveness of foundation models across diverse applications. In this report, we present Atlas, a novel vision foundation model based on the RudolfV approach. Our model was trained on a dataset comprising 1.2 million histopathology whole slide images, collected from two medical institutions: Mayo Clinic and Charit\'e - Universt\"atsmedizin Berlin. Comprehensive evaluations show that Atlas achieves state-of-the-art performance across twenty-one public benchmark datasets, even though it is neither the largest model by parameter count nor by training dataset size.
xCG: Explainable Cell Graphs for Survival Prediction in Non-Small Cell Lung Cancer
Nov 12, 2024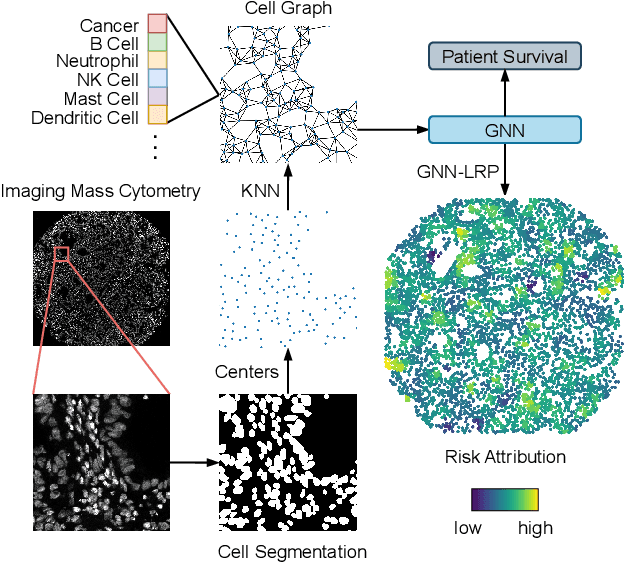
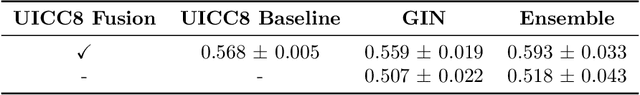
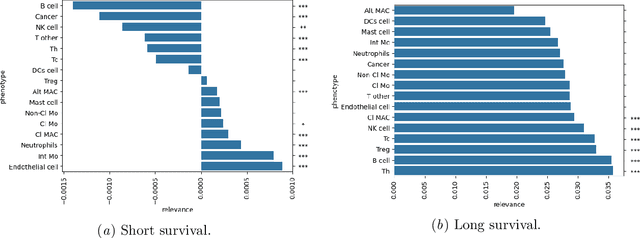
Abstract:Understanding how deep learning models predict oncology patient risk can provide critical insights into disease progression, support clinical decision-making, and pave the way for trustworthy and data-driven precision medicine. Building on recent advances in the spatial modeling of the tumor microenvironment using graph neural networks, we present an explainable cell graph (xCG) approach for survival prediction. We validate our model on a public cohort of imaging mass cytometry (IMC) data for 416 cases of lung adenocarcinoma. We explain survival predictions in terms of known phenotypes on the cell level by computing risk attributions over cell graphs, for which we propose an efficient grid-based layer-wise relevance propagation (LRP) method. Our ablation studies highlight the importance of incorporating the cancer stage and model ensembling to improve the quality of risk estimates. Our xCG method, together with the IMC data, is made publicly available to support further research.
RudolfV: A Foundation Model by Pathologists for Pathologists
Jan 23, 2024



Abstract:Histopathology plays a central role in clinical medicine and biomedical research. While artificial intelligence shows promising results on many pathological tasks, generalization and dealing with rare diseases, where training data is scarce, remains a challenge. Distilling knowledge from unlabeled data into a foundation model before learning from, potentially limited, labeled data provides a viable path to address these challenges. In this work, we extend the state of the art of foundation models for digital pathology whole slide images by semi-automated data curation and incorporating pathologist domain knowledge. Specifically, we combine computational and pathologist domain knowledge (1) to curate a diverse dataset of 103k slides corresponding to 750 million image patches covering data from different fixation, staining, and scanning protocols as well as data from different indications and labs across the EU and US, (2) for grouping semantically similar slides and tissue patches, and (3) to augment the input images during training. We evaluate the resulting model on a set of public and internal benchmarks and show that although our foundation model is trained with an order of magnitude less slides, it performs on par or better than competing models. We expect that scaling our approach to more data and larger models will further increase its performance and capacity to deal with increasingly complex real world tasks in diagnostics and biomedical research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge