Marvin Sextro
MapPFN: Learning Causal Perturbation Maps in Context
Jan 28, 2026Abstract:Planning effective interventions in biological systems requires treatment-effect models that adapt to unseen biological contexts by identifying their specific underlying mechanisms. Yet single-cell perturbation datasets span only a handful of biological contexts, and existing methods cannot leverage new interventional evidence at inference time to adapt beyond their training data. To meta-learn a perturbation effect estimator, we present MapPFN, a prior-data fitted network (PFN) pretrained on synthetic data generated from a prior over causal perturbations. Given a set of experiments, MapPFN uses in-context learning to predict post-perturbation distributions, without gradient-based optimization. Despite being pretrained on in silico gene knockouts alone, MapPFN identifies differentially expressed genes, matching the performance of models trained on real single-cell data. Our code and data are available at https://github.com/marvinsxtr/MapPFN.
xCG: Explainable Cell Graphs for Survival Prediction in Non-Small Cell Lung Cancer
Nov 12, 2024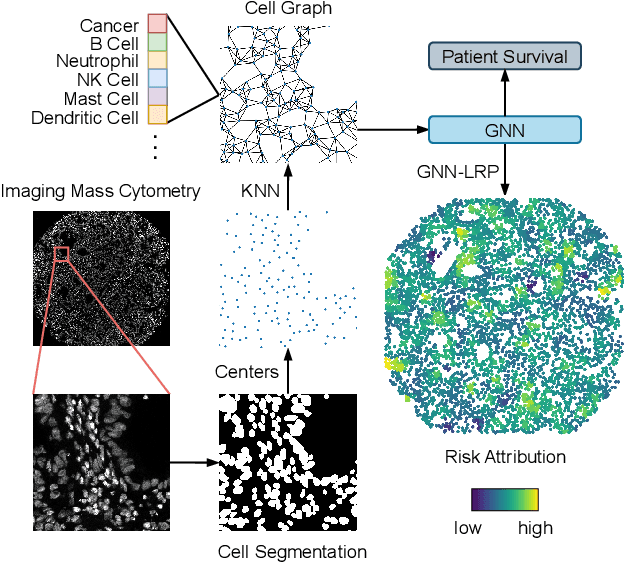
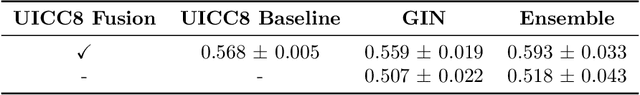
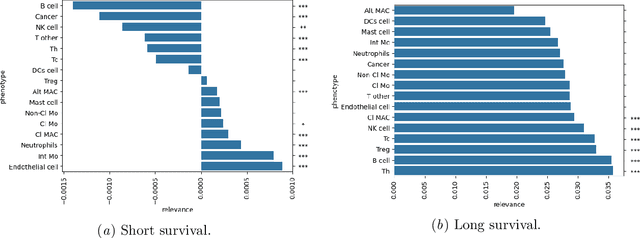
Abstract:Understanding how deep learning models predict oncology patient risk can provide critical insights into disease progression, support clinical decision-making, and pave the way for trustworthy and data-driven precision medicine. Building on recent advances in the spatial modeling of the tumor microenvironment using graph neural networks, we present an explainable cell graph (xCG) approach for survival prediction. We validate our model on a public cohort of imaging mass cytometry (IMC) data for 416 cases of lung adenocarcinoma. We explain survival predictions in terms of known phenotypes on the cell level by computing risk attributions over cell graphs, for which we propose an efficient grid-based layer-wise relevance propagation (LRP) method. Our ablation studies highlight the importance of incorporating the cancer stage and model ensembling to improve the quality of risk estimates. Our xCG method, together with the IMC data, is made publicly available to support further research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge