Jussi Tohka
for the Alzheimerś Disease Neuroimaging Initiative
USFetal: Tools for Fetal Brain Ultrasound Compounding
Jan 11, 2026Abstract:Ultrasound offers a safe, cost-effective, and widely accessible technology for fetal brain imaging, making it especially suitable for routine clinical use. However, it suffers from view-dependent artifacts, operator variability, and a limited field of view, which make interpretation and quantitative evaluation challenging. Ultrasound compounding aims to overcome these limitations by integrating complementary information from multiple 3D acquisitions into a single, coherent volumetric representation. This work provides four main contributions: (1) We present the first systematic categorization of computational strategies for fetal brain ultrasound compounding, including both classical techniques and modern learning-based frameworks. (2) We implement and compare representative methods across four key categories - multi-scale, transformation-based, variational, and deep learning approaches - emphasizing their core principles and practical advantages. (3) Motivated by the lack of full-view, artifact-free ground truth required for supervised learning, we focus on unsupervised and self-supervised strategies and introduce two new deep learning based approaches: a self-supervised compounding framework and an adaptation of unsupervised deep plug-and-play priors for compounding. (4) We conduct a comprehensive evaluation on ten multi-view fetal brain ultrasound datasets, using both expert radiologist scoring and standard quantitative image-quality metrics. We also release the USFetal Compounding Toolbox, publicly available to support benchmarking and future research. Keywords: Ultrasound compounding, fetal brain, deep learning, self-supervised, unsupervised.
Federated Learning for Predicting Mild Cognitive Impairment to Dementia Conversion
Mar 05, 2025Abstract:Dementia is a progressive condition that impairs an individual's cognitive health and daily functioning, with mild cognitive impairment (MCI) often serving as its precursor. The prediction of MCI to dementia conversion has been well studied, but previous studies have almost always focused on traditional Machine Learning (ML) based methods that require sharing sensitive clinical information to train predictive models. This study proposes a privacy-enhancing solution using Federated Learning (FL) to train predictive models for MCI to dementia conversion without sharing sensitive data, leveraging socio demographic and cognitive measures. We simulated and compared two network architectures, Peer to Peer (P2P) and client-server, to enable collaborative learning. Our results demonstrated that FL had comparable predictive performance to centralized ML, and each clinical site showed similar performance without sharing local data. Moreover, the predictive performance of FL models was superior to site specific models trained without collaboration. This work highlights that FL can eliminate the need for data sharing without compromising model efficacy.
Unified Bayesian representation for high-dimensional multi-modal biomedical data for small-sample classification
Nov 11, 2024Abstract:We present BALDUR, a novel Bayesian algorithm designed to deal with multi-modal datasets and small sample sizes in high-dimensional settings while providing explainable solutions. To do so, the proposed model combines within a common latent space the different data views to extract the relevant information to solve the classification task and prune out the irrelevant/redundant features/data views. Furthermore, to provide generalizable solutions in small sample size scenarios, BALDUR efficiently integrates dual kernels over the views with a small sample-to-feature ratio. Finally, its linear nature ensures the explainability of the model outcomes, allowing its use for biomarker identification. This model was tested over two different neurodegeneration datasets, outperforming the state-of-the-art models and detecting features aligned with markers already described in the scientific literature.
Segment Anything for Dendrites from Electron Microscopy
Nov 04, 2024Abstract:Segmentation of cellular structures in electron microscopy (EM) images is fundamental to analyzing the morphology of neurons and glial cells in the healthy and diseased brain tissue. Current neuronal segmentation applications are based on convolutional neural networks (CNNs) and do not effectively capture global relationships within images. Here, we present DendriteSAM, a vision foundation model based on Segment Anything, for interactive and automatic segmentation of dendrites in EM images. The model is trained on high-resolution EM data from healthy rat hippocampus and is tested on diseased rat and human data. Our evaluation results demonstrate better mask quality compared to the original and other fine-tuned models, leveraging the features learned during training. This study introduces the first implementation of vision foundation models in dendrite segmentation, paving the path for computer-assisted diagnosis of neuronal anomalies.
No-Clean-Reference Image Super-Resolution: Application to Electron Microscopy
Jan 26, 2024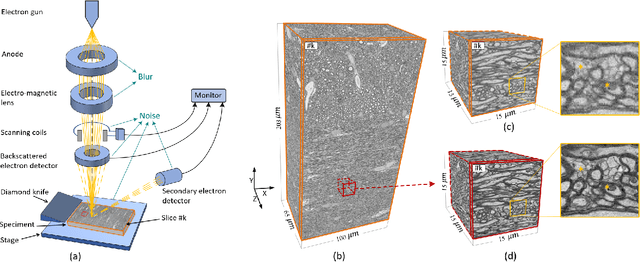


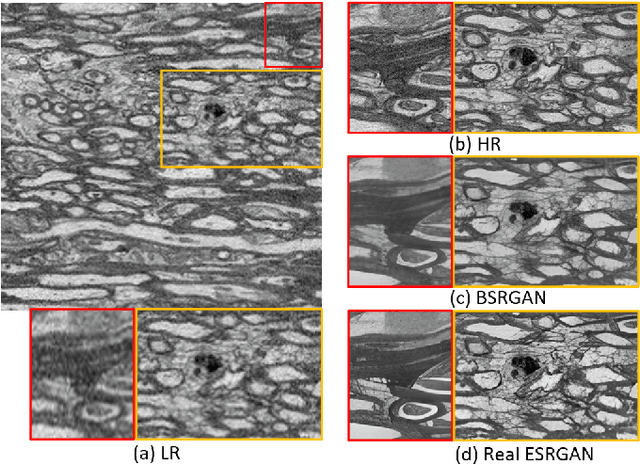
Abstract:The inability to acquire clean high-resolution (HR) electron microscopy (EM) images over a large brain tissue volume hampers many neuroscience studies. To address this challenge, we propose a deep-learning-based image super-resolution (SR) approach to computationally reconstruct clean HR 3D-EM with a large field of view (FoV) from noisy low-resolution (LR) acquisition. Our contributions are I) Investigating training with no-clean references for $\ell_2$ and $\ell_1$ loss functions; II) Introducing a novel network architecture, named EMSR, for enhancing the resolution of LR EM images while reducing inherent noise; and, III) Comparing different training strategies including using acquired LR and HR image pairs, i.e., real pairs with no-clean references contaminated with real corruptions, the pairs of synthetic LR and acquired HR, as well as acquired LR and denoised HR pairs. Experiments with nine brain datasets showed that training with real pairs can produce high-quality super-resolved results, demonstrating the feasibility of training with non-clean references for both loss functions. Additionally, comparable results were observed, both visually and numerically, when employing denoised and noisy references for training. Moreover, utilizing the network trained with synthetically generated LR images from HR counterparts proved effective in yielding satisfactory SR results, even in certain cases, outperforming training with real pairs. The proposed SR network was compared quantitatively and qualitatively with several established SR techniques, showcasing either the superiority or competitiveness of the proposed method in mitigating noise while recovering fine details.
Optimizing Feature Selection for Binary Classification with Noisy Labels: A Genetic Algorithm Approach
Jan 12, 2024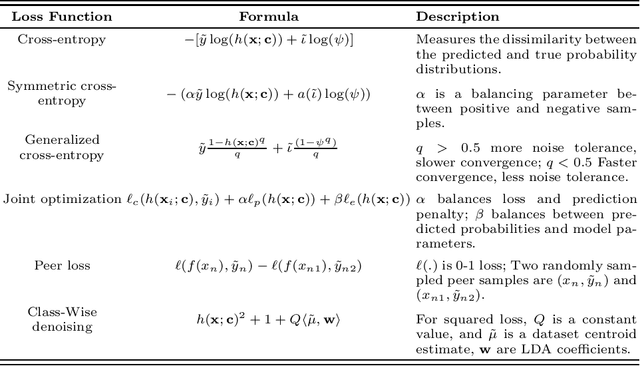
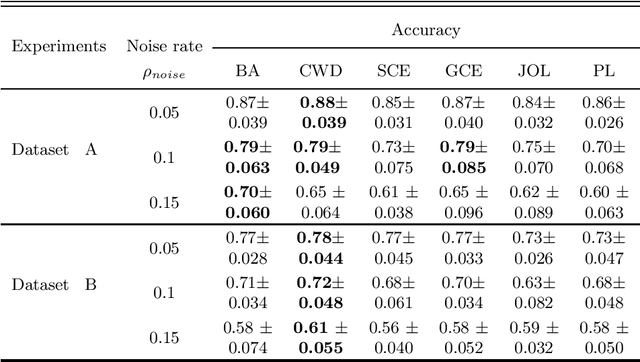
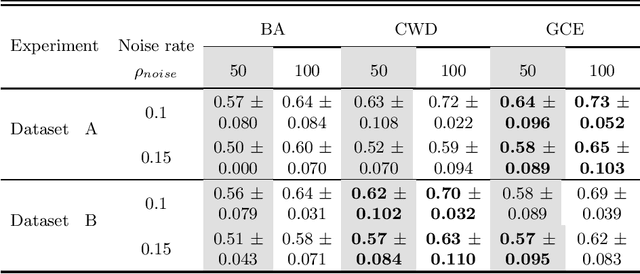
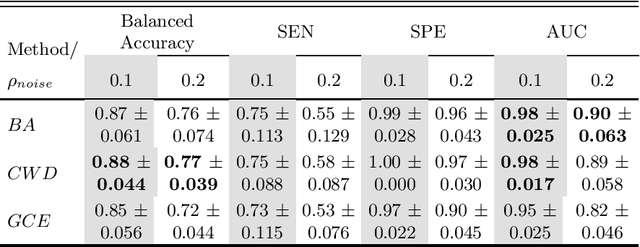
Abstract:Feature selection in noisy label scenarios remains an understudied topic. We propose a novel genetic algorithm-based approach, the Noise-Aware Multi-Objective Feature Selection Genetic Algorithm (NMFS-GA), for selecting optimal feature subsets in binary classification with noisy labels. NMFS-GA offers a unified framework for selecting feature subsets that are both accurate and interpretable. We evaluate NMFS-GA on synthetic datasets with label noise, a Breast Cancer dataset enriched with noisy features, and a real-world ADNI dataset for dementia conversion prediction. Our results indicate that NMFS-GA can effectively select feature subsets that improve the accuracy and interpretability of binary classifiers in scenarios with noisy labels.
Convolutional Neural Networks for Automatic Detection of Intact Adenovirus from TEM Imaging with Debris, Broken and Artefacts Particles
Nov 06, 2023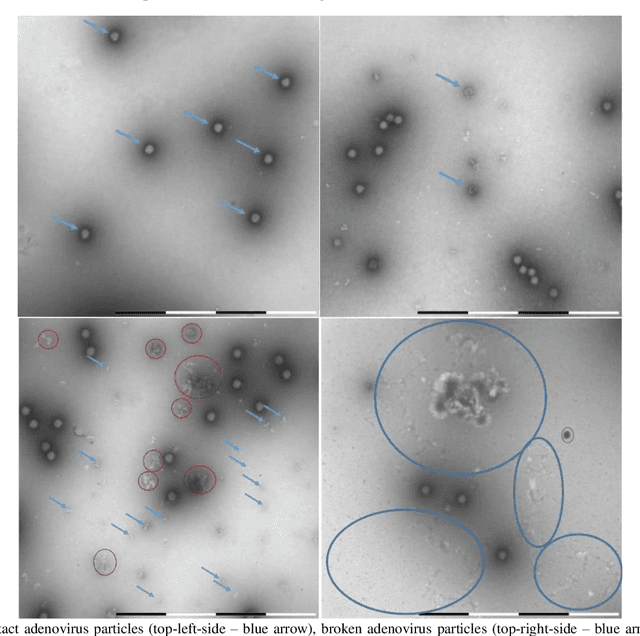
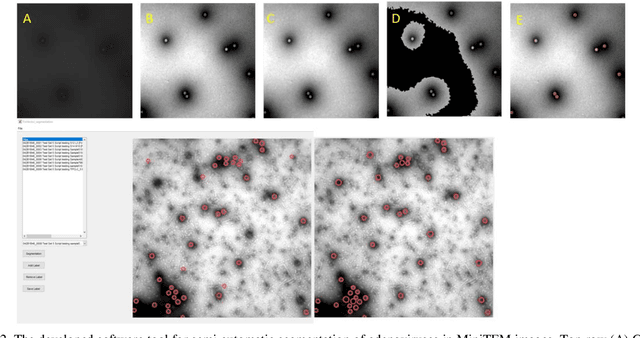
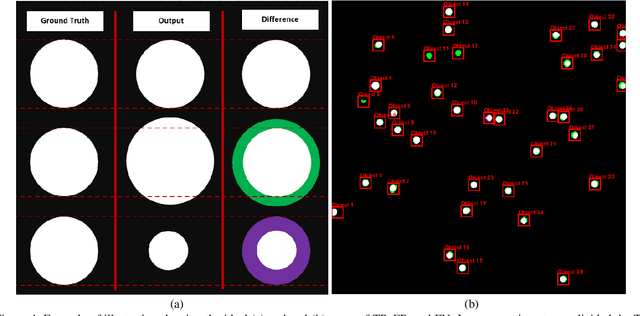
Abstract:Regular monitoring of the primary particles and purity profiles of a drug product during development and manufacturing processes is essential for manufacturers to avoid product variability and contamination. Transmission electron microscopy (TEM) imaging helps manufacturers predict how changes affect particle characteristics and purity for virus-based gene therapy vector products and intermediates. Since intact particles can characterize efficacious products, it is beneficial to automate the detection of intact adenovirus against a non-intact-viral background mixed with debris, broken, and artefact particles. In the presence of such particles, detecting intact adenoviruses becomes more challenging. To overcome the challenge, due to such a presence, we developed a software tool for semi-automatic annotation and segmentation of adenoviruses and a software tool for automatic segmentation and detection of intact adenoviruses in TEM imaging systems. The developed semi-automatic tool exploited conventional image analysis techniques while the automatic tool was built based on convolutional neural networks and image analysis techniques. Our quantitative and qualitative evaluations showed outstanding true positive detection rates compared to false positive and negative rates where adenoviruses were nicely detected without mistaking them for real debris, broken adenoviruses, and/or staining artefacts.
Self-Supervised Super-Resolution Approach for Isotropic Reconstruction of 3D Electron Microscopy Images from Anisotropic Acquisition
Sep 19, 2023Abstract:Three-dimensional electron microscopy (3DEM) is an essential technique to investigate volumetric tissue ultra-structure. Due to technical limitations and high imaging costs, samples are often imaged anisotropically, where resolution in the axial direction ($z$) is lower than in the lateral directions $(x,y)$. This anisotropy 3DEM can hamper subsequent analysis and visualization tasks. To overcome this limitation, we propose a novel deep-learning (DL)-based self-supervised super-resolution approach that computationally reconstructs isotropic 3DEM from the anisotropic acquisition. The proposed DL-based framework is built upon the U-shape architecture incorporating vision-transformer (ViT) blocks, enabling high-capability learning of local and global multi-scale image dependencies. To train the tailored network, we employ a self-supervised approach. Specifically, we generate pairs of anisotropic and isotropic training datasets from the given anisotropic 3DEM data. By feeding the given anisotropic 3DEM dataset in the trained network through our proposed framework, the isotropic 3DEM is obtained. Importantly, this isotropic reconstruction approach relies solely on the given anisotropic 3DEM dataset and does not require pairs of co-registered anisotropic and isotropic 3DEM training datasets. To evaluate the effectiveness of the proposed method, we conducted experiments using three 3DEM datasets acquired from brain. The experimental results demonstrated that our proposed framework could successfully reconstruct isotropic 3DEM from the anisotropic acquisition.
Multi-Objective Genetic Algorithm for Multi-View Feature Selection
May 26, 2023Abstract:Multi-view datasets offer diverse forms of data that can enhance prediction models by providing complementary information. However, the use of multi-view data leads to an increase in high-dimensional data, which poses significant challenges for the prediction models that can lead to poor generalization. Therefore, relevant feature selection from multi-view datasets is important as it not only addresses the poor generalization but also enhances the interpretability of the models. Despite the success of traditional feature selection methods, they have limitations in leveraging intrinsic information across modalities, lacking generalizability, and being tailored to specific classification tasks. We propose a novel genetic algorithm strategy to overcome these limitations of traditional feature selection methods for multi-view data. Our proposed approach, called the multi-view multi-objective feature selection genetic algorithm (MMFS-GA), simultaneously selects the optimal subset of features within a view and between views under a unified framework. The MMFS-GA framework demonstrates superior performance and interpretability for feature selection on multi-view datasets in both binary and multiclass classification tasks. The results of our evaluations on three benchmark datasets, including synthetic and real data, show improvement over the best baseline methods. This work provides a promising solution for multi-view feature selection and opens up new possibilities for further research in multi-view datasets.
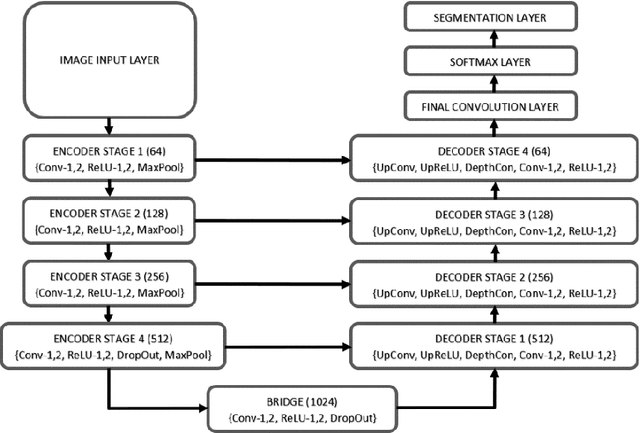
Sauron U-Net: Simple automated redundancy elimination in medical image segmentation via filter pruning
Sep 27, 2022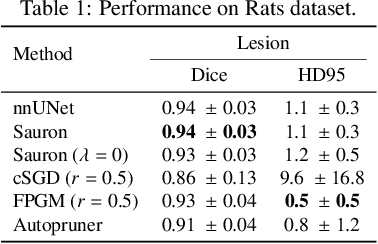
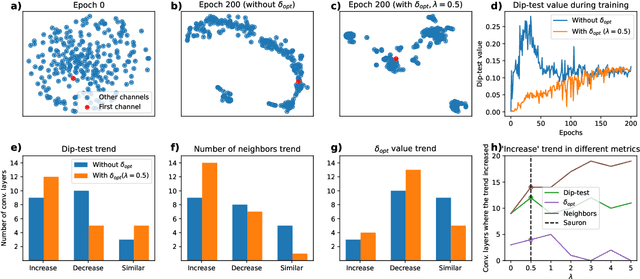
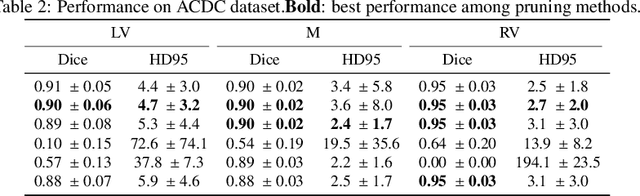
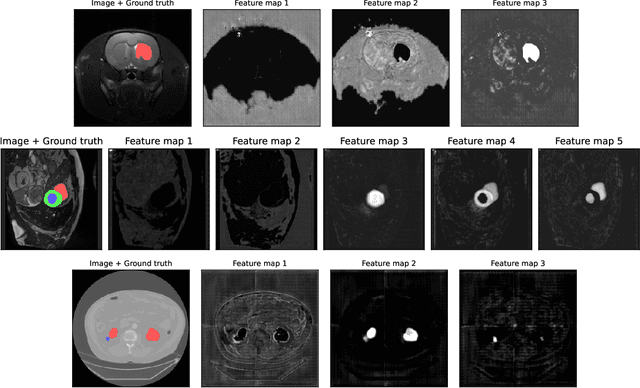
Abstract:We present Sauron, a filter pruning method that eliminates redundant feature maps by discarding the corresponding filters with automatically-adjusted layer-specific thresholds. Furthermore, Sauron minimizes a regularization term that, as we show with various metrics, promotes the formation of feature maps clusters. In contrast to most filter pruning methods, Sauron is single-phase, similarly to typical neural network optimization, requiring fewer hyperparameters and design decisions. Additionally, unlike other cluster-based approaches, our method does not require pre-selecting the number of clusters, which is non-trivial to determine and varies across layers. We evaluated Sauron and three state-of-the-art filter pruning methods on three medical image segmentation tasks. This is an area where filter pruning has received little attention and where it can help building efficient models for medical grade computers that cannot use cloud services due to privacy considerations. Sauron achieved models with higher performance and pruning rate than the competing pruning methods. Additionally, since Sauron removes filters during training, its optimization accelerated over time. Finally, we show that the feature maps of a Sauron-pruned model were highly interpretable. The Sauron code is publicly available at https://github.com/jmlipman/SauronUNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge