Mohammad Khateri
USFetal: Tools for Fetal Brain Ultrasound Compounding
Jan 11, 2026Abstract:Ultrasound offers a safe, cost-effective, and widely accessible technology for fetal brain imaging, making it especially suitable for routine clinical use. However, it suffers from view-dependent artifacts, operator variability, and a limited field of view, which make interpretation and quantitative evaluation challenging. Ultrasound compounding aims to overcome these limitations by integrating complementary information from multiple 3D acquisitions into a single, coherent volumetric representation. This work provides four main contributions: (1) We present the first systematic categorization of computational strategies for fetal brain ultrasound compounding, including both classical techniques and modern learning-based frameworks. (2) We implement and compare representative methods across four key categories - multi-scale, transformation-based, variational, and deep learning approaches - emphasizing their core principles and practical advantages. (3) Motivated by the lack of full-view, artifact-free ground truth required for supervised learning, we focus on unsupervised and self-supervised strategies and introduce two new deep learning based approaches: a self-supervised compounding framework and an adaptation of unsupervised deep plug-and-play priors for compounding. (4) We conduct a comprehensive evaluation on ten multi-view fetal brain ultrasound datasets, using both expert radiologist scoring and standard quantitative image-quality metrics. We also release the USFetal Compounding Toolbox, publicly available to support benchmarking and future research. Keywords: Ultrasound compounding, fetal brain, deep learning, self-supervised, unsupervised.
No-Clean-Reference Image Super-Resolution: Application to Electron Microscopy
Jan 26, 2024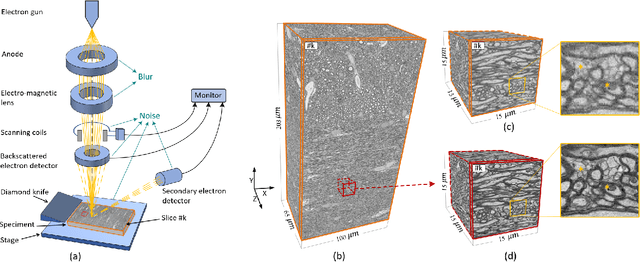


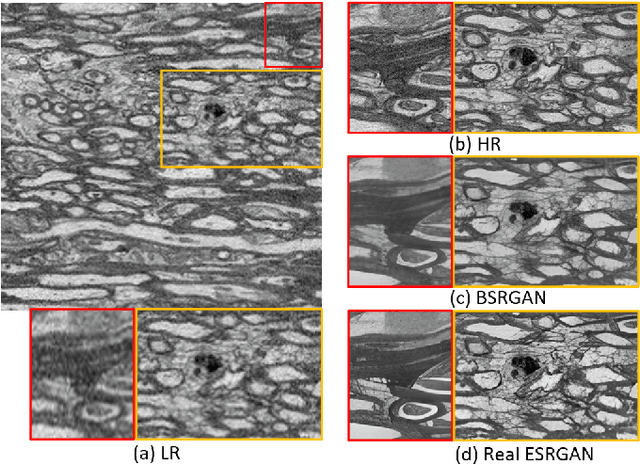
Abstract:The inability to acquire clean high-resolution (HR) electron microscopy (EM) images over a large brain tissue volume hampers many neuroscience studies. To address this challenge, we propose a deep-learning-based image super-resolution (SR) approach to computationally reconstruct clean HR 3D-EM with a large field of view (FoV) from noisy low-resolution (LR) acquisition. Our contributions are I) Investigating training with no-clean references for $\ell_2$ and $\ell_1$ loss functions; II) Introducing a novel network architecture, named EMSR, for enhancing the resolution of LR EM images while reducing inherent noise; and, III) Comparing different training strategies including using acquired LR and HR image pairs, i.e., real pairs with no-clean references contaminated with real corruptions, the pairs of synthetic LR and acquired HR, as well as acquired LR and denoised HR pairs. Experiments with nine brain datasets showed that training with real pairs can produce high-quality super-resolved results, demonstrating the feasibility of training with non-clean references for both loss functions. Additionally, comparable results were observed, both visually and numerically, when employing denoised and noisy references for training. Moreover, utilizing the network trained with synthetically generated LR images from HR counterparts proved effective in yielding satisfactory SR results, even in certain cases, outperforming training with real pairs. The proposed SR network was compared quantitatively and qualitatively with several established SR techniques, showcasing either the superiority or competitiveness of the proposed method in mitigating noise while recovering fine details.
Self-Supervised Super-Resolution Approach for Isotropic Reconstruction of 3D Electron Microscopy Images from Anisotropic Acquisition
Sep 19, 2023Abstract:Three-dimensional electron microscopy (3DEM) is an essential technique to investigate volumetric tissue ultra-structure. Due to technical limitations and high imaging costs, samples are often imaged anisotropically, where resolution in the axial direction ($z$) is lower than in the lateral directions $(x,y)$. This anisotropy 3DEM can hamper subsequent analysis and visualization tasks. To overcome this limitation, we propose a novel deep-learning (DL)-based self-supervised super-resolution approach that computationally reconstructs isotropic 3DEM from the anisotropic acquisition. The proposed DL-based framework is built upon the U-shape architecture incorporating vision-transformer (ViT) blocks, enabling high-capability learning of local and global multi-scale image dependencies. To train the tailored network, we employ a self-supervised approach. Specifically, we generate pairs of anisotropic and isotropic training datasets from the given anisotropic 3DEM data. By feeding the given anisotropic 3DEM dataset in the trained network through our proposed framework, the isotropic 3DEM is obtained. Importantly, this isotropic reconstruction approach relies solely on the given anisotropic 3DEM dataset and does not require pairs of co-registered anisotropic and isotropic 3DEM training datasets. To evaluate the effectiveness of the proposed method, we conducted experiments using three 3DEM datasets acquired from brain. The experimental results demonstrated that our proposed framework could successfully reconstruct isotropic 3DEM from the anisotropic acquisition.
Adversarial Distortion Learning for Medical Image Denoising
Apr 29, 2022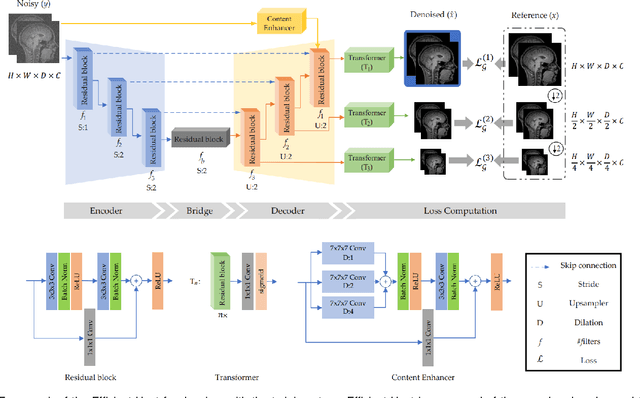
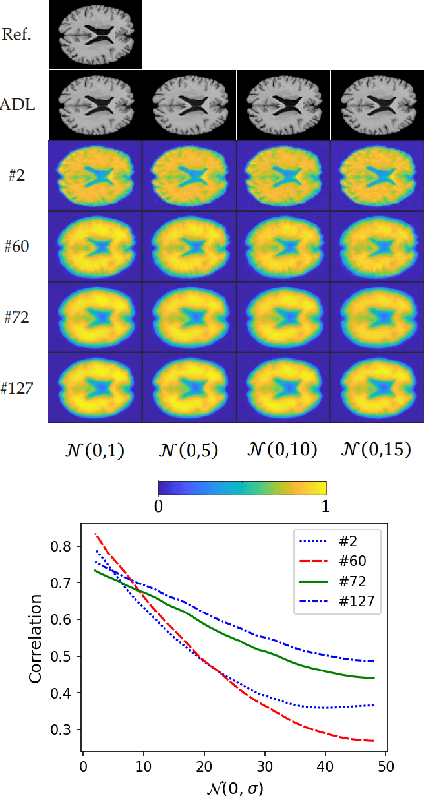
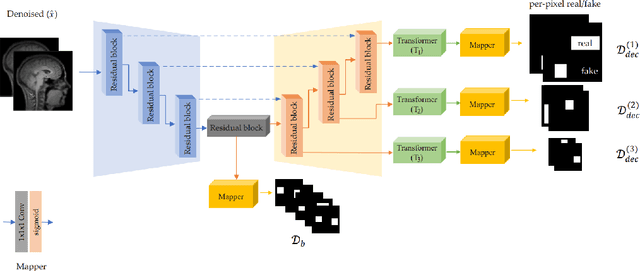
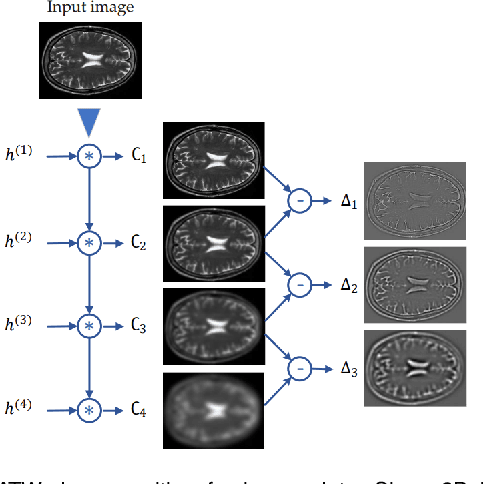
Abstract:We present a novel adversarial distortion learning (ADL) for denoising two- and three-dimensional (2D/3D) biomedical image data. The proposed ADL consists of two auto-encoders: a denoiser and a discriminator. The denoiser removes noise from input data and the discriminator compares the denoised result to its noise-free counterpart. This process is repeated until the discriminator cannot differentiate the denoised data from the reference. Both the denoiser and the discriminator are built upon a proposed auto-encoder called Efficient-Unet. Efficient-Unet has a light architecture that uses the residual blocks and a novel pyramidal approach in the backbone to efficiently extract and re-use feature maps. During training, the textural information and contrast are controlled by two novel loss functions. The architecture of Efficient-Unet allows generalizing the proposed method to any sort of biomedical data. The 2D version of our network was trained on ImageNet and tested on biomedical datasets whose distribution is completely different from ImageNet; so, there is no need for re-training. Experimental results carried out on magnetic resonance imaging (MRI), dermatoscopy, electron microscopy and X-ray datasets show that the proposed method achieved the best on each benchmark. Our implementation and pre-trained models are available at https://github.com/mogvision/ADL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge