Vandad Imani
Optimizing Feature Selection for Binary Classification with Noisy Labels: A Genetic Algorithm Approach
Jan 12, 2024
Abstract:Feature selection in noisy label scenarios remains an understudied topic. We propose a novel genetic algorithm-based approach, the Noise-Aware Multi-Objective Feature Selection Genetic Algorithm (NMFS-GA), for selecting optimal feature subsets in binary classification with noisy labels. NMFS-GA offers a unified framework for selecting feature subsets that are both accurate and interpretable. We evaluate NMFS-GA on synthetic datasets with label noise, a Breast Cancer dataset enriched with noisy features, and a real-world ADNI dataset for dementia conversion prediction. Our results indicate that NMFS-GA can effectively select feature subsets that improve the accuracy and interpretability of binary classifiers in scenarios with noisy labels.
Multi-Objective Genetic Algorithm for Multi-View Feature Selection
May 26, 2023Abstract:Multi-view datasets offer diverse forms of data that can enhance prediction models by providing complementary information. However, the use of multi-view data leads to an increase in high-dimensional data, which poses significant challenges for the prediction models that can lead to poor generalization. Therefore, relevant feature selection from multi-view datasets is important as it not only addresses the poor generalization but also enhances the interpretability of the models. Despite the success of traditional feature selection methods, they have limitations in leveraging intrinsic information across modalities, lacking generalizability, and being tailored to specific classification tasks. We propose a novel genetic algorithm strategy to overcome these limitations of traditional feature selection methods for multi-view data. Our proposed approach, called the multi-view multi-objective feature selection genetic algorithm (MMFS-GA), simultaneously selects the optimal subset of features within a view and between views under a unified framework. The MMFS-GA framework demonstrates superior performance and interpretability for feature selection on multi-view datasets in both binary and multiclass classification tasks. The results of our evaluations on three benchmark datasets, including synthetic and real data, show improvement over the best baseline methods. This work provides a promising solution for multi-view feature selection and opens up new possibilities for further research in multi-view datasets.
Multi-task longitudinal forecasting with missing values on Alzheimer's Disease
Jan 13, 2022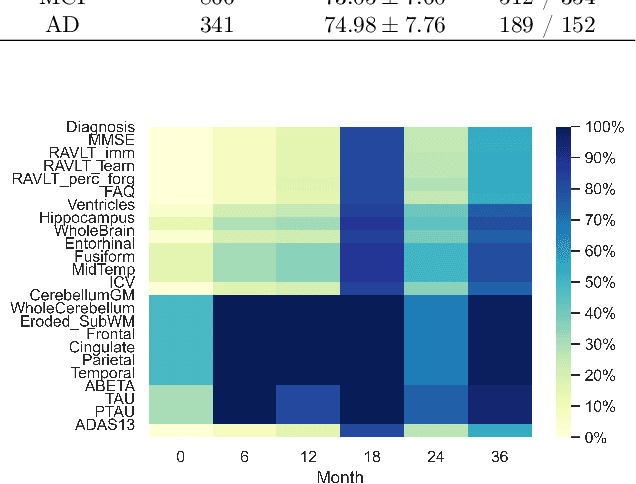



Abstract:Machine learning techniques typically applied to dementia forecasting lack in their capabilities to jointly learn several tasks, handle time dependent heterogeneous data and missing values. In this paper, we propose a framework using the recently presented SSHIBA model for jointly learning different tasks on longitudinal data with missing values. The method uses Bayesian variational inference to impute missing values and combine information of several views. This way, we can combine different data-views from different time-points in a common latent space and learn the relations between each time-point while simultaneously modelling and predicting several output variables. We apply this model to predict together diagnosis, ventricle volume, and clinical scores in dementia. The results demonstrate that SSHIBA is capable of learning a good imputation of the missing values and outperforming the baselines while simultaneously predicting three different tasks.
Comparison of single and multitask learning for predicting cognitive decline based on MRI data
Sep 21, 2021
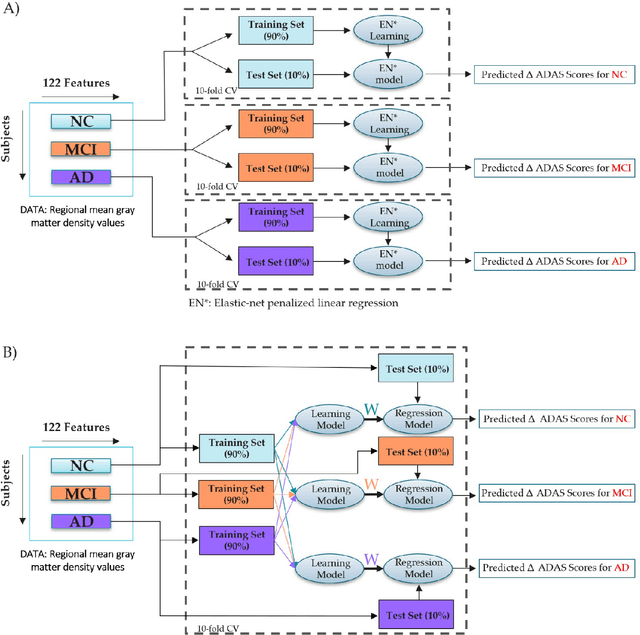


Abstract:The Alzheimer's Disease Assessment Scale-Cognitive subscale (ADAS-Cog) is a neuropsychological tool that has been designed to assess the severity of cognitive symptoms of dementia. Personalized prediction of the changes in ADAS-Cog scores could help in timing therapeutic interventions in dementia and at-risk populations. In the present work, we compared single and multitask learning approaches to predict the changes in ADAS-Cog scores based on T1-weighted anatomical magnetic resonance imaging (MRI). In contrast to most machine learning-based prediction methods ADAS-Cog changes, we stratified the subjects based on their baseline diagnoses and evaluated the prediction performances in each group. Our experiments indicated a positive relationship between the predicted and observed ADAS-Cog score changes in each diagnostic group, suggesting that T1-weighted MRI has a predictive value for evaluating cognitive decline in the entire AD continuum. We further studied whether correction of the differences in the magnetic field strength of MRI would improve the ADAS-Cog score prediction. The partial least square-based domain adaptation slightly improved the prediction performance, but the improvement was marginal. In summary, this study demonstrated that ADAS-Cog change could be, to some extent, predicted based on anatomical MRI. Based on this study, the recommended method for learning the predictive models is a single-task regularized linear regression due to its simplicity and good performance. It appears important to combine the training data across all subject groups for the most effective predictive models.
Transfer Learning in Magnetic Resonance Brain Imaging: a Systematic Review
Feb 02, 2021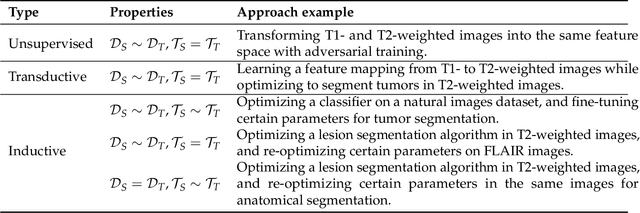
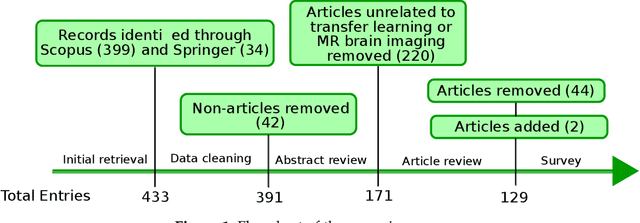

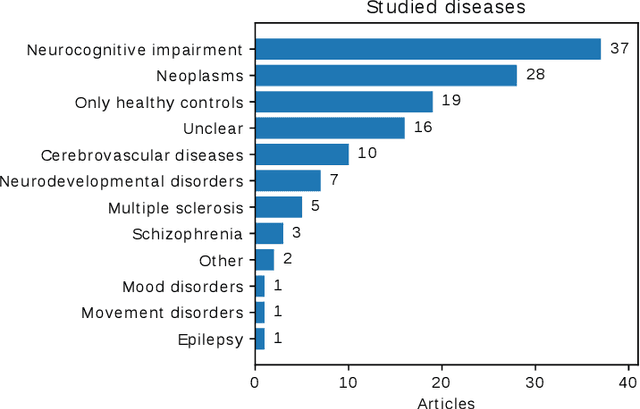
Abstract:Transfer learning refers to machine learning techniques that focus on acquiring knowledge from related tasks to improve generalization in the tasks of interest. In MRI, transfer learning is important for developing strategies that address the variation in MR images. Additionally, transfer learning is beneficial to re-utilize machine learning models that were trained to solve related tasks to the task of interest. Our goal is to identify research directions, gaps of knowledge, applications, and widely used strategies among the transfer learning approaches applied in MR brain imaging. We performed a systematic literature search for articles that applied transfer learning to MR brain imaging. We screened 433 studies and we categorized and extracted relevant information, including task type, application, and machine learning methods. Furthermore, we closely examined brain MRI-specific transfer learning approaches and other methods that tackled privacy, unseen target domains, and unlabeled data. We found 129 articles that applied transfer learning to brain MRI tasks. The most frequent applications were dementia related classification tasks and brain tumor segmentation. A majority of articles utilized transfer learning on convolutional neural networks (CNNs). Only few approaches were clearly brain MRI specific, considered privacy issues, unseen target domains or unlabeled data. We proposed a new categorization to group specific, widely-used approaches. There is an increasing interest in transfer learning within brain MRI. Public datasets have contributed to the popularity of Alzheimer's diagnostics/prognostics and tumor segmentation. Likewise, the availability of pretrained CNNs has promoted their utilization. Finally, the majority of the surveyed studies did not examine in detail the interpretation of their strategies after applying transfer learning, and did not compare to other approaches.
Predicting intelligence based on cortical WM/GM contrast, cortical thickness and volumetry
Sep 09, 2019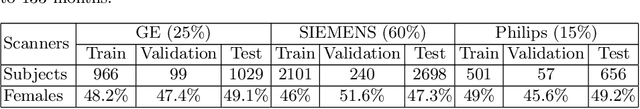
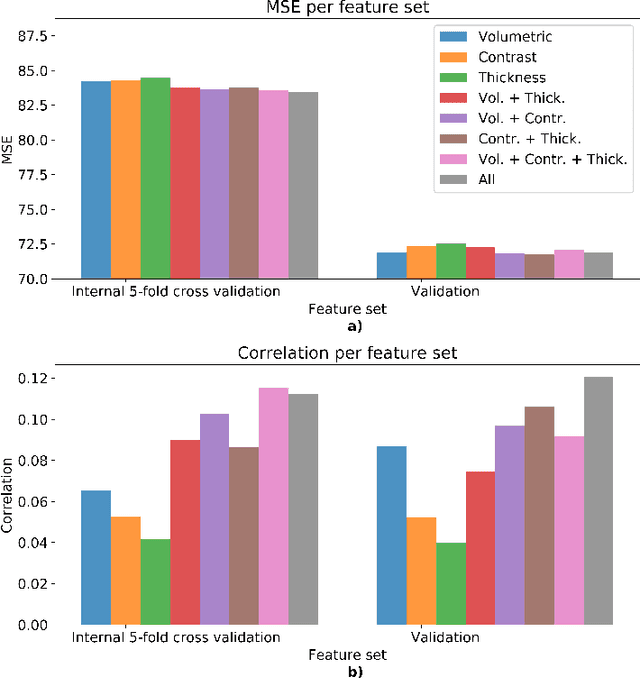
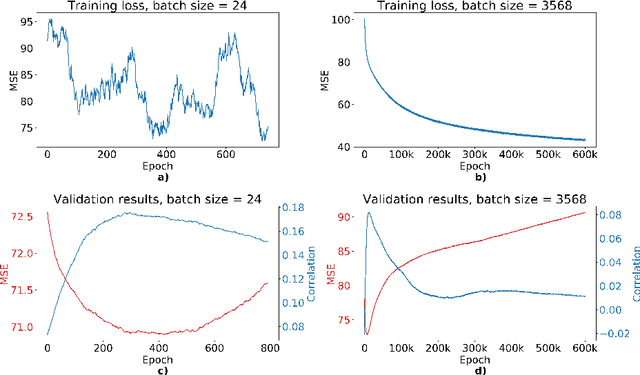
Abstract:We propose a four-layer fully-connected neural network (FNN) for predicting fluid intelligence scores from T1-weighted MR images for the ABCD-challenge. In addition to the volumes of brain structures, the FNN uses cortical WM/GM contrast and cortical thickness at 78 cortical regions. These last two measurements were derived from the T1-weighted MR images using cortical surfaces produced by the CIVET pipeline. The age and gender of the subjects and the scanner manufacturer are also used as features for the learning algorithm. This yielded 283 features provided to the FNN with two hidden layers of 20 and 15 nodes. The method was applied to the data from the ABCD study. Trained with a training set of 3736 subjects, the proposed method achieved a MSE of 71.596 and a correlation of 0.151 in the validation set of 415 subjects. For the final submission, the model was trained with 3568 subjects and it achieved a MSE of 94.0270 in the test set comprised of 4383 subjects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge