Eija Jokitalo
Segment Anything for Dendrites from Electron Microscopy
Nov 04, 2024Abstract:Segmentation of cellular structures in electron microscopy (EM) images is fundamental to analyzing the morphology of neurons and glial cells in the healthy and diseased brain tissue. Current neuronal segmentation applications are based on convolutional neural networks (CNNs) and do not effectively capture global relationships within images. Here, we present DendriteSAM, a vision foundation model based on Segment Anything, for interactive and automatic segmentation of dendrites in EM images. The model is trained on high-resolution EM data from healthy rat hippocampus and is tested on diseased rat and human data. Our evaluation results demonstrate better mask quality compared to the original and other fine-tuned models, leveraging the features learned during training. This study introduces the first implementation of vision foundation models in dendrite segmentation, paving the path for computer-assisted diagnosis of neuronal anomalies.
A Hitchhiker`s Guide through the Bio-image Analysis Software Universe
Apr 15, 2022
Abstract:Modern research in the life sciences is unthinkable without computational methods for extracting, quantifying and visualizing information derived from biological microscopy imaging data. In the past decade, we observed a dramatic increase in available software packages for these purposes. As it is increasingly difficult to keep track of the number of available image analysis platforms, tool collections, components and emerging technologies, we provide a conservative overview of software we use in daily routine and give insights into emerging new tools. We give guidance on which aspects to consider when choosing the right platform, including aspects such as image data type, skills of the team, infrastructure and community at the institute and availability of time and budget.
gACSON software for automated segmentation and morphology analyses of myelinated axons in 3D electron microscopy
Dec 13, 2021



Abstract:Background and Objective: Advances in electron microscopy (EM) now allow three-dimensional (3D) imaging of hundreds of micrometers of tissue with nanometer-scale resolution, providing new opportunities to study the ultrastructure of the brain. In this work, we introduce a freely available gACSON software for visualization, segmentation, assessment, and morphology analysis of myelinated axons in 3D-EM volumes of brain tissue samples. Methods: The gACSON software is equipped with a graphical user interface (GUI). It automatically segments the intra-axonal space of myelinated axons and their corresponding myelin sheaths and allows manual segmentation, proofreading, and interactive correction of the segmented components. gACSON analyzes the morphology of myelinated axons, such as axonal diameter, axonal eccentricity, myelin thickness, or g-ratio. Results: We illustrate the use of gACSON by segmenting and analyzing myelinated axons in six 3D-EM volumes of rat somatosensory cortex after sham surgery or traumatic brain injury (TBI). Our results suggest that the equivalent diameter of myelinated axons in somatisensory cortex was decreased in TBI animals five months after the injury. Conclusions: Our results indicate that gACSON is a valuable tool for visualization, segmentation, assessment, and morphology analysis of myelinated axons in 3D-EM volumes. gACSON is freely available at https://github.com/AndreaBehan/g-ACSON under the MIT license.
Code-free development and deployment of deep segmentation models for digital pathology
Nov 16, 2021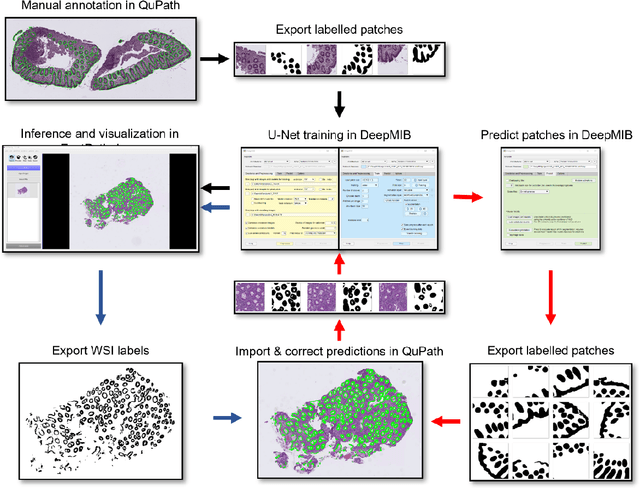
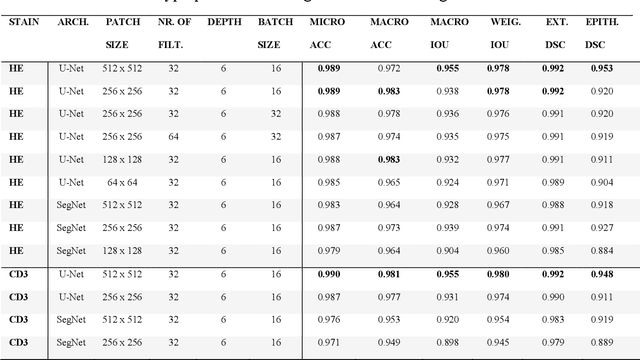
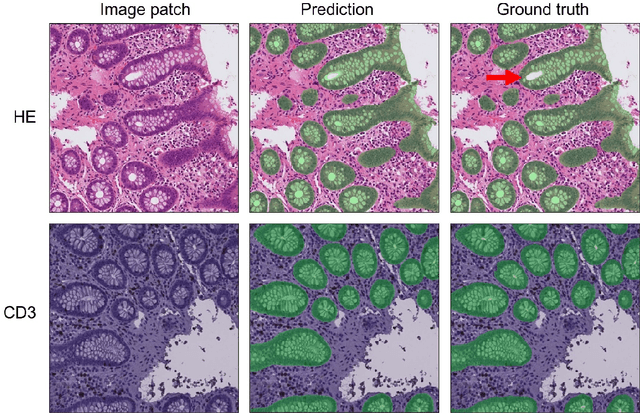
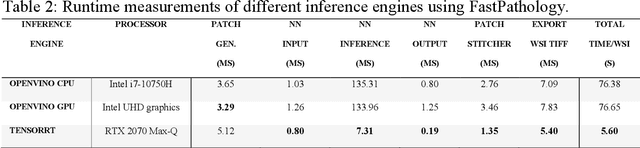
Abstract:Application of deep learning on histopathological whole slide images (WSIs) holds promise of improving diagnostic efficiency and reproducibility but is largely dependent on the ability to write computer code or purchase commercial solutions. We present a code-free pipeline utilizing free-to-use, open-source software (QuPath, DeepMIB, and FastPathology) for creating and deploying deep learning-based segmentation models for computational pathology. We demonstrate the pipeline on a use case of separating epithelium from stroma in colonic mucosa. A dataset of 251 annotated WSIs, comprising 140 hematoxylin-eosin (HE)-stained and 111 CD3 immunostained colon biopsy WSIs, were developed through active learning using the pipeline. On a hold-out test set of 36 HE and 21 CD3-stained WSIs a mean intersection over union score of 96.6% and 95.3% was achieved on epithelium segmentation. We demonstrate pathologist-level segmentation accuracy and clinical acceptable runtime performance and show that pathologists without programming experience can create near state-of-the-art segmentation solutions for histopathological WSIs using only free-to-use software. The study further demonstrates the strength of open-source solutions in its ability to create generalizable, open pipelines, of which trained models and predictions can seamlessly be exported in open formats and thereby used in external solutions. All scripts, trained models, a video tutorial, and the full dataset of 251 WSIs with ~31k epithelium annotations are made openly available at https://github.com/andreped/NoCodeSeg to accelerate research in the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge