Michael Doube
How many radiographs are needed to re-train a deep learning system for object detection?
Oct 17, 2022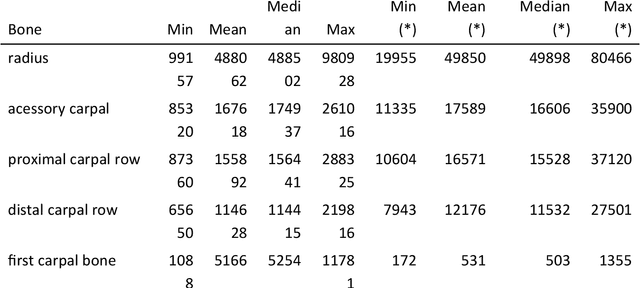

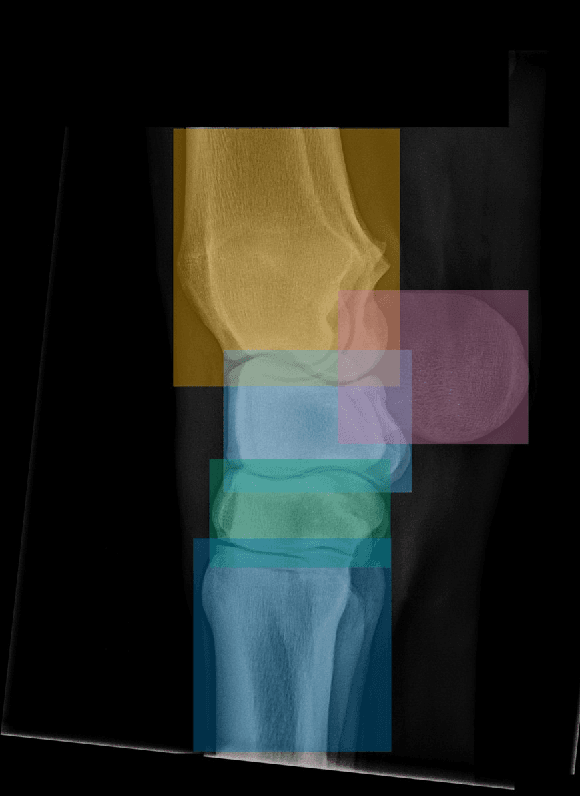
Abstract:Background: Object detection in radiograph computer vision has largely benefited from progress in deep convolutional neural networks and can, for example, annotate a radiograph with a box around a knee joint or intervertebral disc. Is deep learning capable of detect small (less than 1% of the image) in radiographs? And how many radiographs do we need use when re-training a deep learning model? Methods: We annotated 396 radiographs of left and right carpi dorsal 75 medial to palmarolateral oblique (DMPLO) projection with the location of radius, proximal row of carpal bones, distal row of carpal bones, accessory carpal bone, first carpal bone (if present), and metacarpus (metacarpal II, III, and IV). The radiographs and respective annotations were splited into sets that were used to leave-one-out cross-validation of models created using transfer learn from YOLOv5s. Results: Models trained using 96 radiographs or more achieved precision, recall and mAP above 0.95, including for the first carpal bone, when trained for 32 epochs. The best model needed the double of epochs to learn to detect the first carpal bone compared with the other bones. Conclusions: Free and open source state of the art object detection models based on deep learning can be re-trained for radiograph computer vision applications with 100 radiographs and achieved precision, recall and mAP above 0.95.
Equine radiograph classification using deep convolutional neural networks
Apr 29, 2022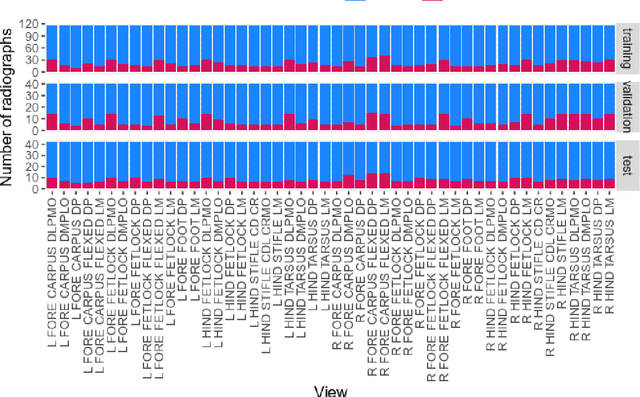
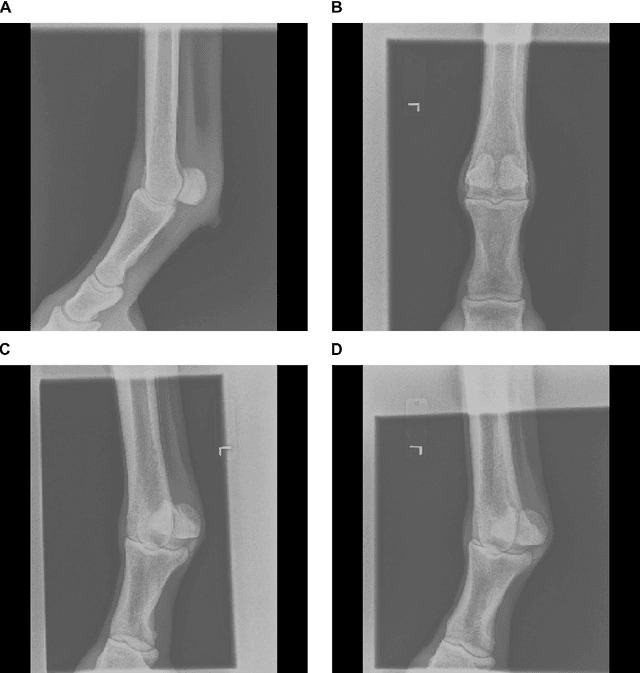
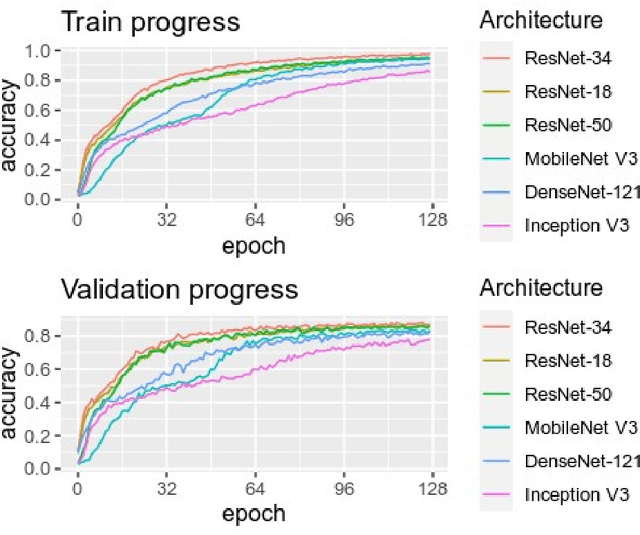
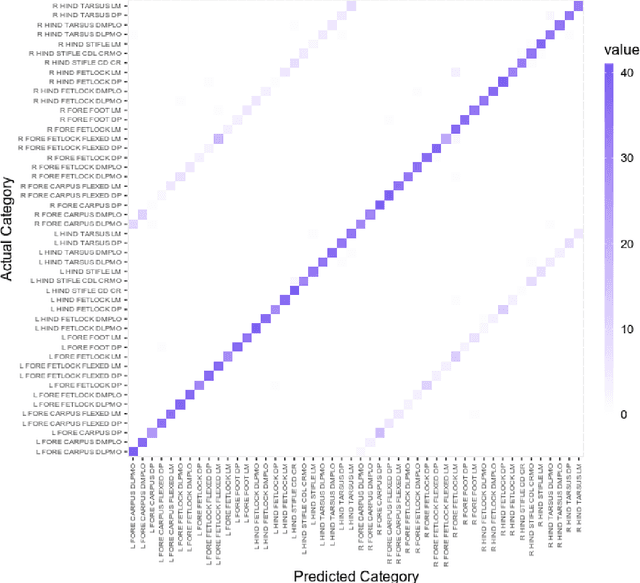
Abstract:Purpose: To assess the capability of deep convolutional neural networks to classify anatomical location and projection from a series of 48 standard views of racehorse limbs. Materials and Methods: 9504 equine pre-import radiographs were used to train, validate, and test six deep learning architectures available as part of the open source machine learning framework PyTorch. Results: ResNet-34 achieved a top-1 accuracy of 0.8408 and the majority (88%) of misclassification was because of wrong laterality. Class activation maps indicated that joint morphology drove the model decision. Conclusion: Deep convolutional neural networks are capable of classifying equine pre-import radiographs into the 48 standard views including moderate discrimination of laterality independent of side marker presence.
A Hitchhiker`s Guide through the Bio-image Analysis Software Universe
Apr 15, 2022
Abstract:Modern research in the life sciences is unthinkable without computational methods for extracting, quantifying and visualizing information derived from biological microscopy imaging data. In the past decade, we observed a dramatic increase in available software packages for these purposes. As it is increasingly difficult to keep track of the number of available image analysis platforms, tool collections, components and emerging technologies, we provide a conservative overview of software we use in daily routine and give insights into emerging new tools. We give guidance on which aspects to consider when choosing the right platform, including aspects such as image data type, skills of the team, infrastructure and community at the institute and availability of time and budget.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge