Artem Shatillo
Sauron U-Net: Simple automated redundancy elimination in medical image segmentation via filter pruning
Sep 27, 2022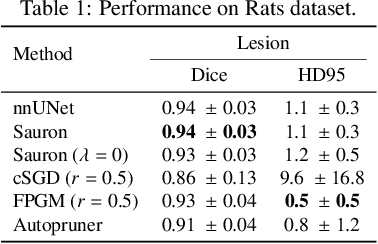
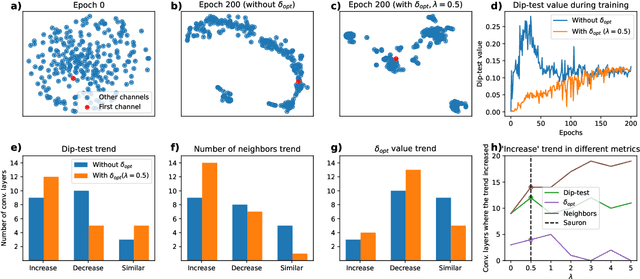
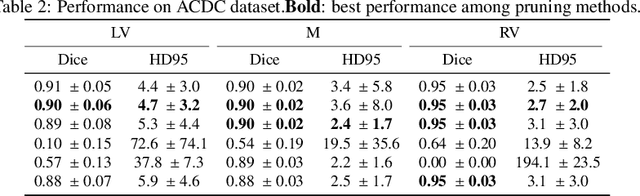
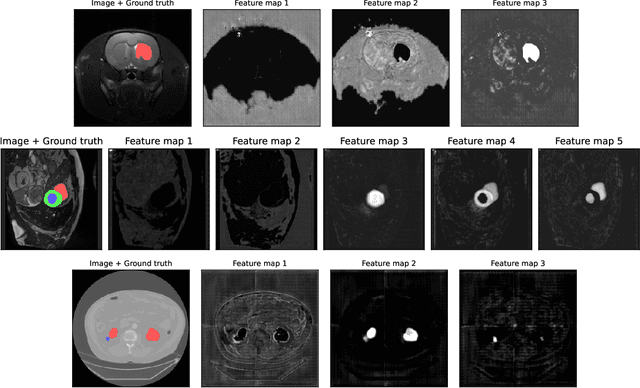
Abstract:We present Sauron, a filter pruning method that eliminates redundant feature maps by discarding the corresponding filters with automatically-adjusted layer-specific thresholds. Furthermore, Sauron minimizes a regularization term that, as we show with various metrics, promotes the formation of feature maps clusters. In contrast to most filter pruning methods, Sauron is single-phase, similarly to typical neural network optimization, requiring fewer hyperparameters and design decisions. Additionally, unlike other cluster-based approaches, our method does not require pre-selecting the number of clusters, which is non-trivial to determine and varies across layers. We evaluated Sauron and three state-of-the-art filter pruning methods on three medical image segmentation tasks. This is an area where filter pruning has received little attention and where it can help building efficient models for medical grade computers that cannot use cloud services due to privacy considerations. Sauron achieved models with higher performance and pruning rate than the competing pruning methods. Additionally, since Sauron removes filters during training, its optimization accelerated over time. Finally, we show that the feature maps of a Sauron-pruned model were highly interpretable. The Sauron code is publicly available at https://github.com/jmlipman/SauronUNet.
Automatic hemisphere segmentation in rodent MRI with lesions
Aug 04, 2021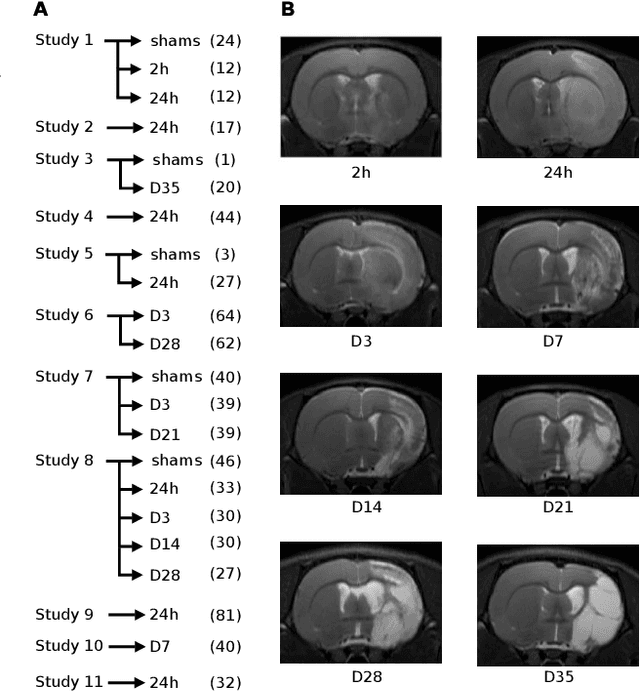
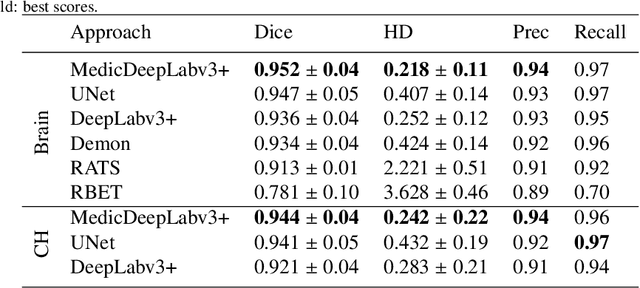
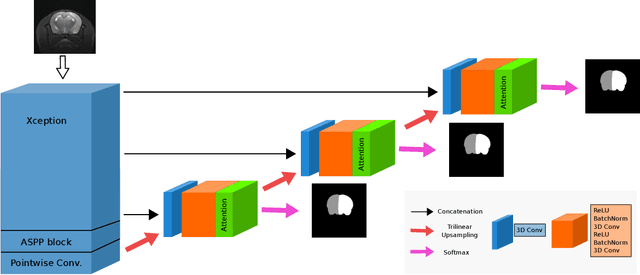
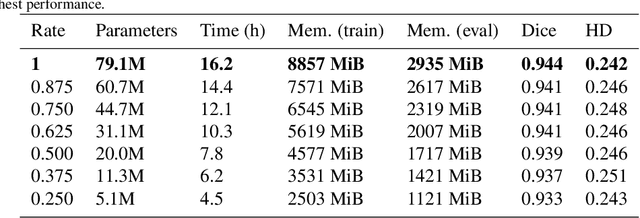
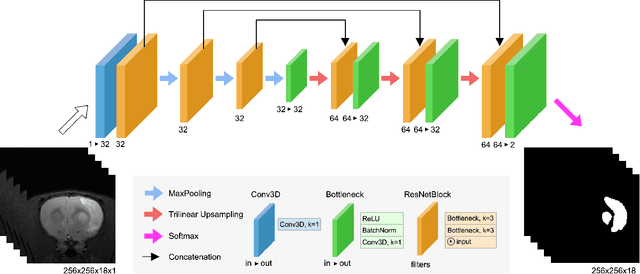
Abstract:We present MedicDeepLabv3+, a convolutional neural network that is the first completely automatic method to segment brain hemispheres in magnetic resonance (MR) images of rodents with lesions. MedicDeepLabv3+ improves the state-of-the-art DeepLabv3+ with an advanced decoder, incorporating spatial attention layers and additional skip connections that, as we show in our experiments, lead to more precise segmentations. MedicDeepLabv3+ requires no MR image preprocessing, such as bias-field correction or registration to a template, produces segmentations in less than a second, and its GPU memory requirements can be adjusted based on the available resources. Using a large dataset of 723 MR rat brain images, we evaluated our MedicDeepLabv3+, two state-of-the-art convolutional neural networks (DeepLabv3+, UNet) and three approaches that were specifically designed for skull-stripping rodent MR images (Demon, RATS and RBET). In our experiments, MedicDeepLabv3+ outperformed the other methods, yielding an average Dice coefficient of 0.952 and 0.944 in the brain and contralateral hemisphere regions. Additionally, we show that despite limiting the GPU memory and the training data to only three images, our MedicDeepLabv3+ also provided satisfactory segmentations. In conclusion, our method, publicly available at https://github.com/jmlipman/MedicDeepLabv3Plus, yielded excellent results in multiple scenarios, demonstrating its capability to reduce human workload in rodent neuroimaging studies.
RatLesNetv2: A Fully Convolutional Network for Rodent Brain Lesion Segmentation
Jan 24, 2020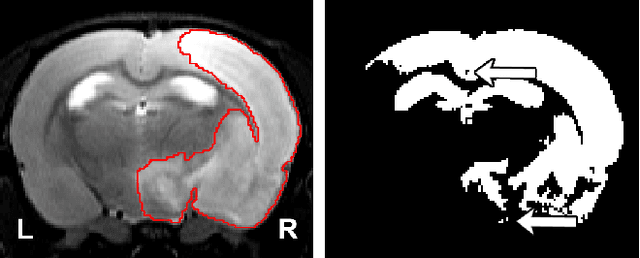

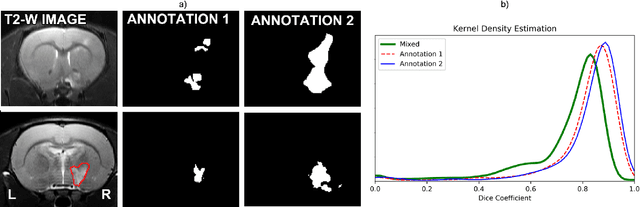
Abstract:Segmentation of rodent brain lesions on magnetic resonance images (MRIs) is a time-consuming task with high inter- and intra-operator variability due to its subjective nature. We present a three-dimensional fully convolutional neural network (ConvNet) named RatLesNetv2 for segmenting rodent brain lesions. We compare its performance with other ConvNets on an unusually large and heterogeneous data set composed by 916 T2-weighted rat brain scans at nine different lesion stages. RatLesNetv2 obtained similar to higher Dice coefficients than the other ConvNets and it produced much more realistic and compact segmentations with notably less holes and lower Hausdorff distance. RatLesNetv2-derived segmentations also exceeded inter-rater agreement Dice coefficients. Additionally, we show that training on disparate ground truths leads to significantly different segmentations, and we study RatLesNetv2 generalization capability when optimizing for training sets of different sizes. RatLesNetv2 is publicly available at https://github.com/jmlipman/RatLesNetv2.
Automatic Rodent Brain MRI Lesion Segmentation with Fully Convolutional Networks
Aug 23, 2019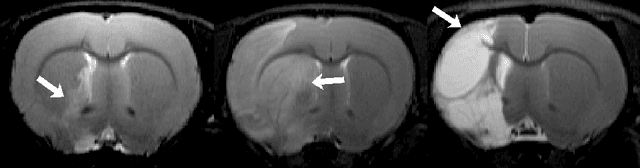
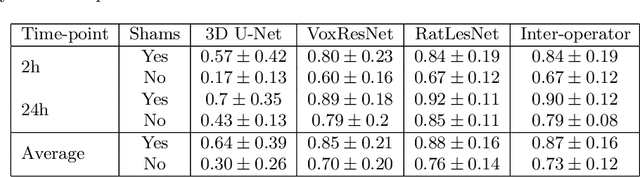
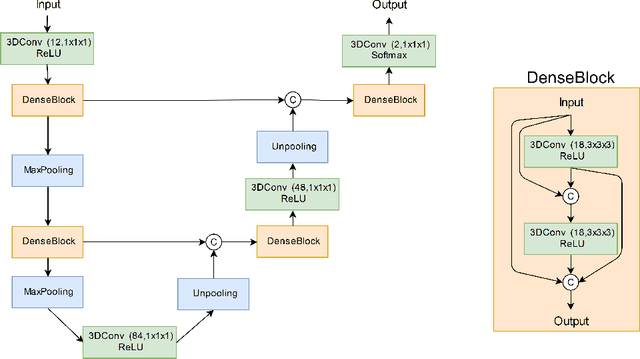
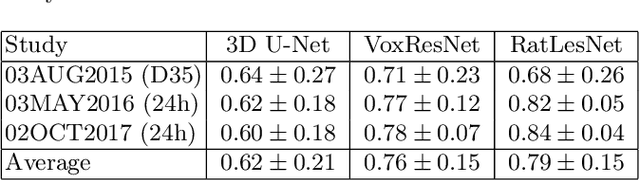
Abstract:Manual segmentation of rodent brain lesions from magnetic resonance images (MRIs) is an arduous, time-consuming and subjective task that is highly important in pre-clinical research. Several automatic methods have been developed for different human brain MRI segmentation, but little research has targeted automatic rodent lesion segmentation. The existing tools for performing automatic lesion segmentation in rodents are constrained by strict assumptions about the data. Deep learning has been successfully used for medical image segmentation. However, there has not been any deep learning approach specifically designed for tackling rodent brain lesion segmentation. In this work, we propose a novel Fully Convolutional Network (FCN), RatLesNet, for the aforementioned task. Our dataset consists of 131 T2-weighted rat brain scans from 4 different studies in which ischemic stroke was induced by transient middle cerebral artery occlusion. We compare our method with two other 3D FCNs originally developed for anatomical segmentation (VoxResNet and 3D-U-Net) with 5-fold cross-validation on a single study and a generalization test, where the training was done on a single study and testing on three remaining studies. The labels generated by our method were quantitatively and qualitatively better than the predictions of the compared methods. The average Dice coefficient achieved in the 5-fold cross-validation experiment with the proposed approach was 0.88, between 3.7% and 38% higher than the compared architectures. The presented architecture also outperformed the other FCNs at generalizing on different studies, achieving the average Dice coefficient of 0.79.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge