Julius Vetter
Effortless, Simulation-Efficient Bayesian Inference using Tabular Foundation Models
Apr 24, 2025Abstract:Simulation-based inference (SBI) offers a flexible and general approach to performing Bayesian inference: In SBI, a neural network is trained on synthetic data simulated from a model and used to rapidly infer posterior distributions for observed data. A key goal for SBI is to achieve accurate inference with as few simulations as possible, especially for expensive simulators. In this work, we address this challenge by repurposing recent probabilistic foundation models for tabular data: We show how tabular foundation models -- specifically TabPFN -- can be used as pre-trained autoregressive conditional density estimators for SBI. We propose Neural Posterior Estimation with Prior-data Fitted Networks (NPE-PF) and show that it is competitive with current SBI approaches in terms of accuracy for both benchmark tasks and two complex scientific inverse problems. Crucially, it often substantially outperforms them in terms of simulation efficiency, sometimes requiring orders of magnitude fewer simulations. NPE-PF eliminates the need for inference network selection, training, and hyperparameter tuning. We also show that it exhibits superior robustness to model misspecification and can be scaled to simulation budgets that exceed the context size limit of TabPFN. NPE-PF provides a new direction for SBI, where training-free, general-purpose inference models offer efficient, easy-to-use, and flexible solutions for a wide range of stochastic inverse problems.
sbi reloaded: a toolkit for simulation-based inference workflows
Nov 26, 2024
Abstract:Scientists and engineers use simulators to model empirically observed phenomena. However, tuning the parameters of a simulator to ensure its outputs match observed data presents a significant challenge. Simulation-based inference (SBI) addresses this by enabling Bayesian inference for simulators, identifying parameters that match observed data and align with prior knowledge. Unlike traditional Bayesian inference, SBI only needs access to simulations from the model and does not require evaluations of the likelihood-function. In addition, SBI algorithms do not require gradients through the simulator, allow for massive parallelization of simulations, and can perform inference for different observations without further simulations or training, thereby amortizing inference. Over the past years, we have developed, maintained, and extended $\texttt{sbi}$, a PyTorch-based package that implements Bayesian SBI algorithms based on neural networks. The $\texttt{sbi}$ toolkit implements a wide range of inference methods, neural network architectures, sampling methods, and diagnostic tools. In addition, it provides well-tested default settings but also offers flexibility to fully customize every step of the simulation-based inference workflow. Taken together, the $\texttt{sbi}$ toolkit enables scientists and engineers to apply state-of-the-art SBI methods to black-box simulators, opening up new possibilities for aligning simulations with empirically observed data.
A Practical Guide to Statistical Distances for Evaluating Generative Models in Science
Mar 19, 2024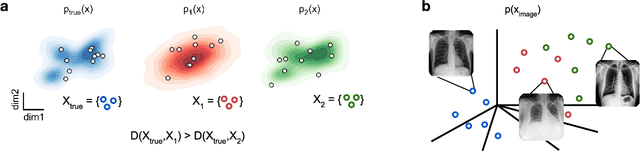
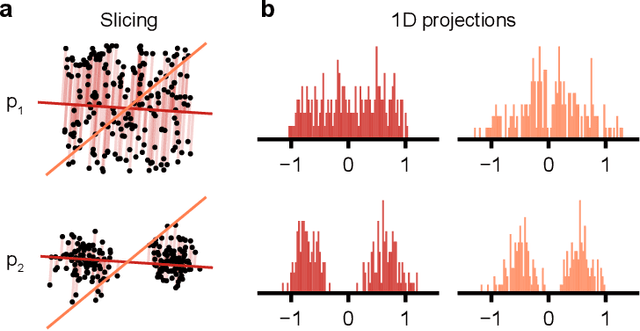
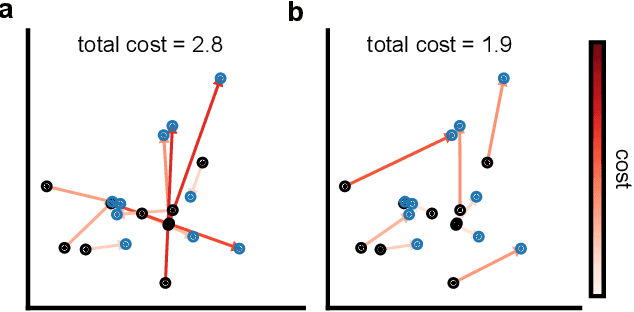
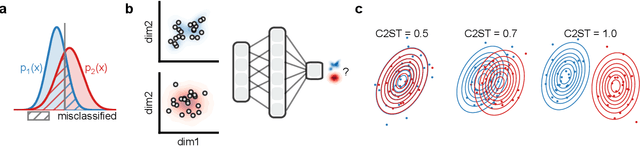
Abstract:Generative models are invaluable in many fields of science because of their ability to capture high-dimensional and complicated distributions, such as photo-realistic images, protein structures, and connectomes. How do we evaluate the samples these models generate? This work aims to provide an accessible entry point to understanding popular notions of statistical distances, requiring only foundational knowledge in mathematics and statistics. We focus on four commonly used notions of statistical distances representing different methodologies: Using low-dimensional projections (Sliced-Wasserstein; SW), obtaining a distance using classifiers (Classifier Two-Sample Tests; C2ST), using embeddings through kernels (Maximum Mean Discrepancy; MMD), or neural networks (Fr\'echet Inception Distance; FID). We highlight the intuition behind each distance and explain their merits, scalability, complexity, and pitfalls. To demonstrate how these distances are used in practice, we evaluate generative models from different scientific domains, namely a model of decision making and a model generating medical images. We showcase that distinct distances can give different results on similar data. Through this guide, we aim to help researchers to use, interpret, and evaluate statistical distances for generative models in science.
Sourcerer: Sample-based Maximum Entropy Source Distribution Estimation
Feb 12, 2024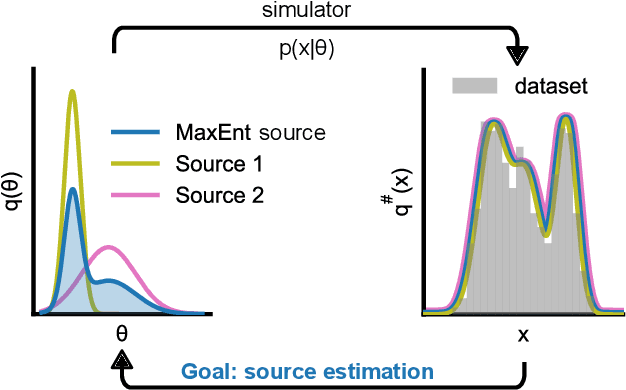
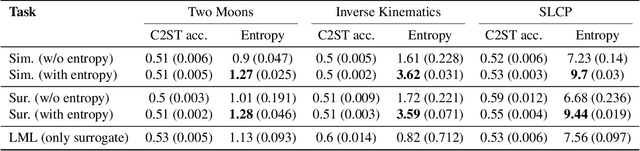
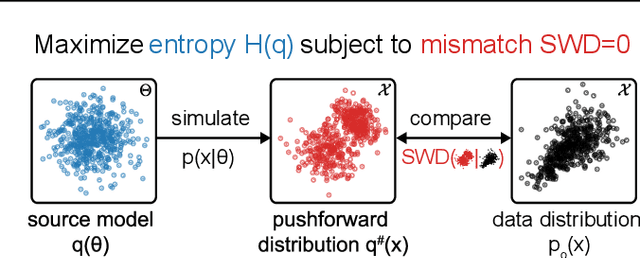
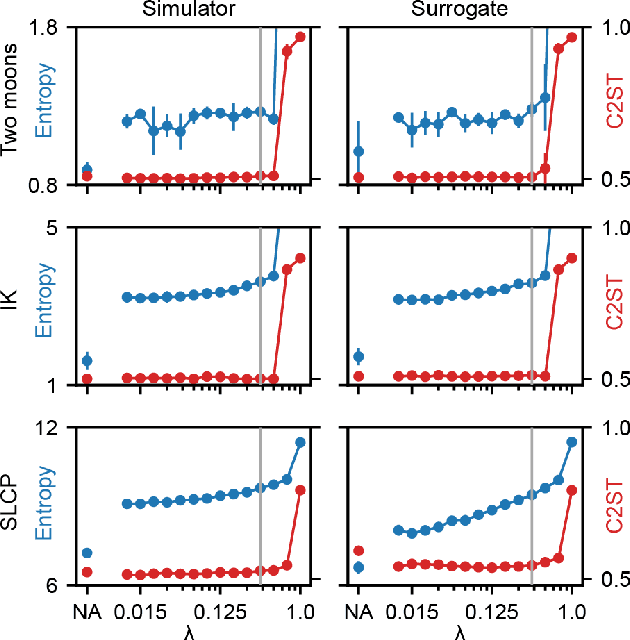
Abstract:Scientific modeling applications often require estimating a distribution of parameters consistent with a dataset of observations - an inference task also known as source distribution estimation. This problem can be ill-posed, however, since many different source distributions might produce the same distribution of data-consistent simulations. To make a principled choice among many equally valid sources, we propose an approach which targets the maximum entropy distribution, i.e., prioritizes retaining as much uncertainty as possible. Our method is purely sample-based - leveraging the Sliced-Wasserstein distance to measure the discrepancy between the dataset and simulations - and thus suitable for simulators with intractable likelihoods. We benchmark our method on several tasks, and show that it can recover source distributions with substantially higher entropy without sacrificing the fidelity of the simulations. Finally, to demonstrate the utility of our approach, we infer source distributions for parameters of the Hodgkin-Huxley neuron model from experimental datasets with thousands of measurements. In summary, we propose a principled framework for inferring unique source distributions of scientific simulator parameters while retaining as much uncertainty as possible.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge