Jakob H Macke
Inferring stochastic low-rank recurrent neural networks from neural data
Jun 24, 2024Abstract:A central aim in computational neuroscience is to relate the activity of large populations of neurons to an underlying dynamical system. Models of these neural dynamics should ideally be both interpretable and fit the observed data well. Low-rank recurrent neural networks (RNNs) exhibit such interpretability by having tractable dynamics. However, it is unclear how to best fit low-rank RNNs to data consisting of noisy observations of an underlying stochastic system. Here, we propose to fit stochastic low-rank RNNs with variational sequential Monte Carlo methods. We validate our method on several datasets consisting of both continuous and spiking neural data, where we obtain lower dimensional latent dynamics than current state of the art methods. Additionally, for low-rank models with piecewise linear nonlinearities, we show how to efficiently identify all fixed points in polynomial rather than exponential cost in the number of units, making analysis of the inferred dynamics tractable for large RNNs. Our method both elucidates the dynamical systems underlying experimental recordings and provides a generative model whose trajectories match observed trial-to-trial variability.
A Practical Guide to Statistical Distances for Evaluating Generative Models in Science
Mar 19, 2024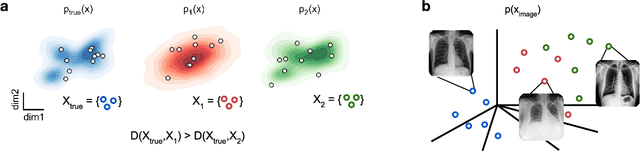
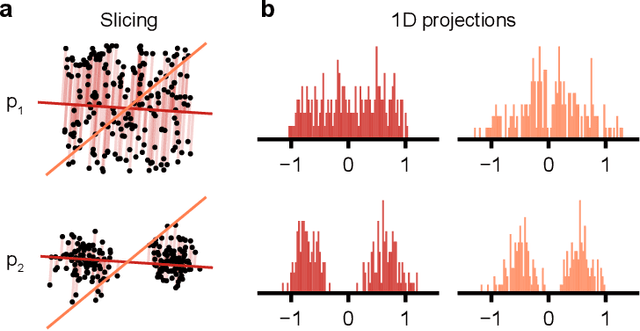
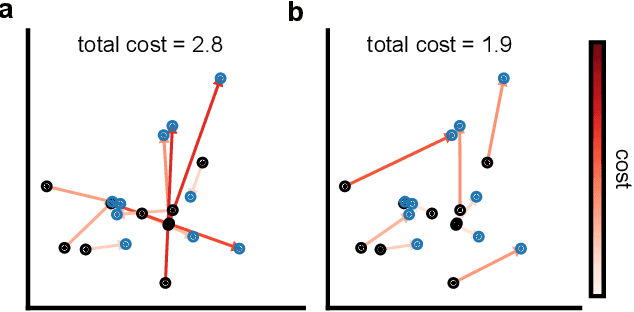
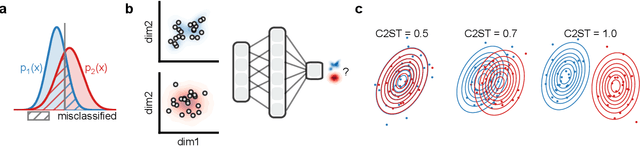
Abstract:Generative models are invaluable in many fields of science because of their ability to capture high-dimensional and complicated distributions, such as photo-realistic images, protein structures, and connectomes. How do we evaluate the samples these models generate? This work aims to provide an accessible entry point to understanding popular notions of statistical distances, requiring only foundational knowledge in mathematics and statistics. We focus on four commonly used notions of statistical distances representing different methodologies: Using low-dimensional projections (Sliced-Wasserstein; SW), obtaining a distance using classifiers (Classifier Two-Sample Tests; C2ST), using embeddings through kernels (Maximum Mean Discrepancy; MMD), or neural networks (Fr\'echet Inception Distance; FID). We highlight the intuition behind each distance and explain their merits, scalability, complexity, and pitfalls. To demonstrate how these distances are used in practice, we evaluate generative models from different scientific domains, namely a model of decision making and a model generating medical images. We showcase that distinct distances can give different results on similar data. Through this guide, we aim to help researchers to use, interpret, and evaluate statistical distances for generative models in science.
Truncated proposals for scalable and hassle-free simulation-based inference
Oct 10, 2022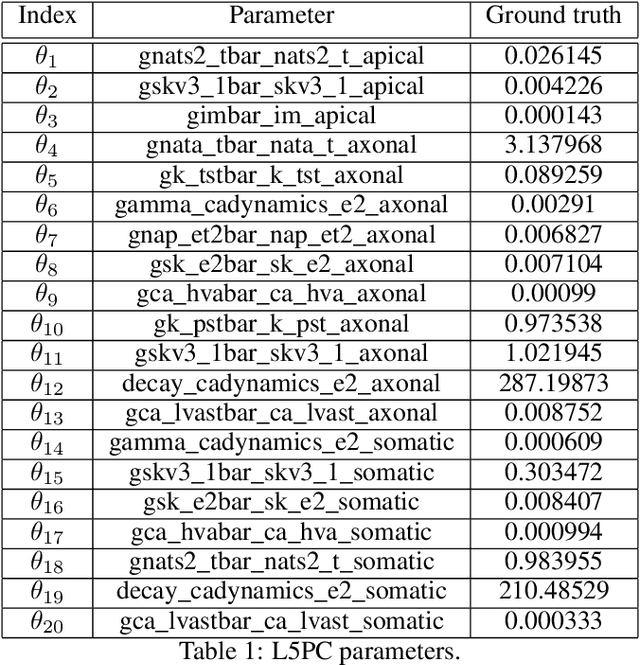
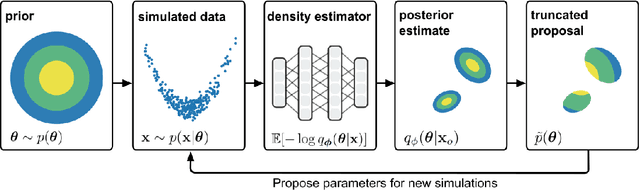
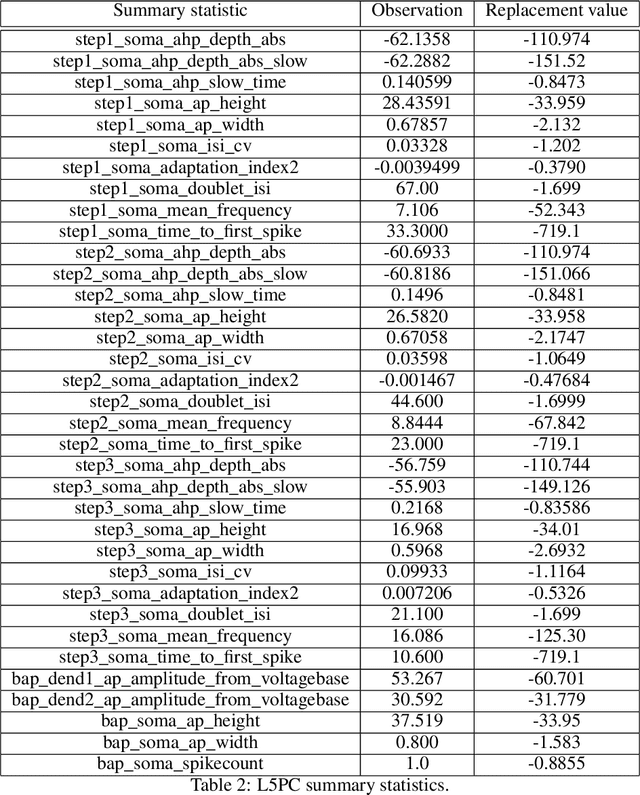
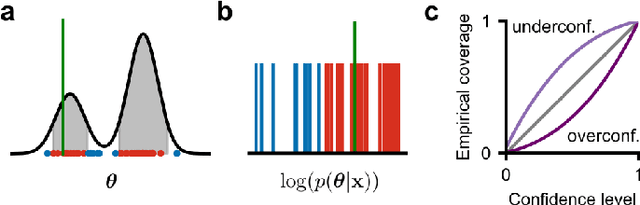
Abstract:Simulation-based inference (SBI) solves statistical inverse problems by repeatedly running a stochastic simulator and inferring posterior distributions from model-simulations. To improve simulation efficiency, several inference methods take a sequential approach and iteratively adapt the proposal distributions from which model simulations are generated. However, many of these sequential methods are difficult to use in practice, both because the resulting optimisation problems can be challenging and efficient diagnostic tools are lacking. To overcome these issues, we present Truncated Sequential Neural Posterior Estimation (TSNPE). TSNPE performs sequential inference with truncated proposals, sidestepping the optimisation issues of alternative approaches. In addition, TSNPE allows to efficiently perform coverage tests that can scale to complex models with many parameters. We demonstrate that TSNPE performs on par with previous methods on established benchmark tasks. We then apply TSNPE to two challenging problems from neuroscience and show that TSNPE can successfully obtain the posterior distributions, whereas previous methods fail. Overall, our results demonstrate that TSNPE is an efficient, accurate, and robust inference method that can scale to challenging scientific models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge