Julien Martinelli
IRS
In-Context Multi-Objective Optimization
Dec 11, 2025Abstract:Balancing competing objectives is omnipresent across disciplines, from drug design to autonomous systems. Multi-objective Bayesian optimization is a promising solution for such expensive, black-box problems: it fits probabilistic surrogates and selects new designs via an acquisition function that balances exploration and exploitation. In practice, it requires tailored choices of surrogate and acquisition that rarely transfer to the next problem, is myopic when multi-step planning is often required, and adds refitting overhead, particularly in parallel or time-sensitive loops. We present TAMO, a fully amortized, universal policy for multi-objective black-box optimization. TAMO uses a transformer architecture that operates across varying input and objective dimensions, enabling pretraining on diverse corpora and transfer to new problems without retraining: at test time, the pretrained model proposes the next design with a single forward pass. We pretrain the policy with reinforcement learning to maximize cumulative hypervolume improvement over full trajectories, conditioning on the entire query history to approximate the Pareto frontier. Across synthetic benchmarks and real tasks, TAMO produces fast proposals, reducing proposal time by 50-1000x versus alternatives while matching or improving Pareto quality under tight evaluation budgets. These results show that transformers can perform multi-objective optimization entirely in-context, eliminating per-task surrogate fitting and acquisition engineering, and open a path to foundation-style, plug-and-play optimizers for scientific discovery workflows.
Robust and Computation-Aware Gaussian Processes
May 27, 2025Abstract:Gaussian processes (GPs) are widely used for regression and optimization tasks such as Bayesian optimization (BO) due to their expressiveness and principled uncertainty estimates. However, in settings with large datasets corrupted by outliers, standard GPs and their sparse approximations struggle with computational tractability and robustness. We introduce Robust Computation-aware Gaussian Process (RCaGP), a novel GP model that jointly addresses these challenges by combining a principled treatment of approximation-induced uncertainty with robust generalized Bayesian updating. The key insight is that robustness and approximation-awareness are not orthogonal but intertwined: approximations can exacerbate the impact of outliers, and mitigating one without the other is insufficient. Unlike previous work that focuses narrowly on either robustness or approximation quality, RCaGP combines both in a principled and scalable framework, thus effectively managing both outliers and computational uncertainties introduced by approximations such as low-rank matrix multiplications. Our model ensures more conservative and reliable uncertainty estimates, a property we rigorously demonstrate. Additionally, we establish a robustness property and show that the mean function is key to preserving it, motivating a tailored model selection scheme for robust mean functions. Empirical results confirm that solving these challenges jointly leads to superior performance across both clean and outlier-contaminated settings, both on regression and high-throughput Bayesian optimization benchmarks.
Challenges in interpretability of additive models
Apr 14, 2025Abstract:We review generalized additive models as a type of ``transparent'' model that has recently seen renewed interest in the deep learning community as neural additive models. We highlight multiple types of nonidentifiability in this model class and discuss challenges in interpretability, arguing for restraint when claiming ``interpretability'' or ``suitability for safety-critical applications'' of such models.
PABBO: Preferential Amortized Black-Box Optimization
Mar 02, 2025Abstract:Preferential Bayesian Optimization (PBO) is a sample-efficient method to learn latent user utilities from preferential feedback over a pair of designs. It relies on a statistical surrogate model for the latent function, usually a Gaussian process, and an acquisition strategy to select the next candidate pair to get user feedback on. Due to the non-conjugacy of the associated likelihood, every PBO step requires a significant amount of computations with various approximate inference techniques. This computational overhead is incompatible with the way humans interact with computers, hindering the use of PBO in real-world cases. Building on the recent advances of amortized BO, we propose to circumvent this issue by fully amortizing PBO, meta-learning both the surrogate and the acquisition function. Our method comprises a novel transformer neural process architecture, trained using reinforcement learning and tailored auxiliary losses. On a benchmark composed of synthetic and real-world datasets, our method is several orders of magnitude faster than the usual Gaussian process-based strategies and often outperforms them in accuracy.
Proxy-informed Bayesian transfer learning with unknown sources
Nov 05, 2024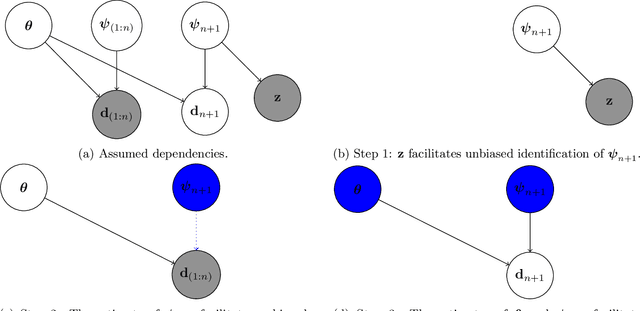
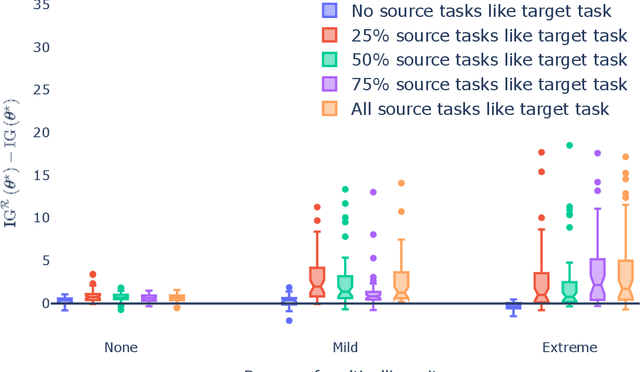
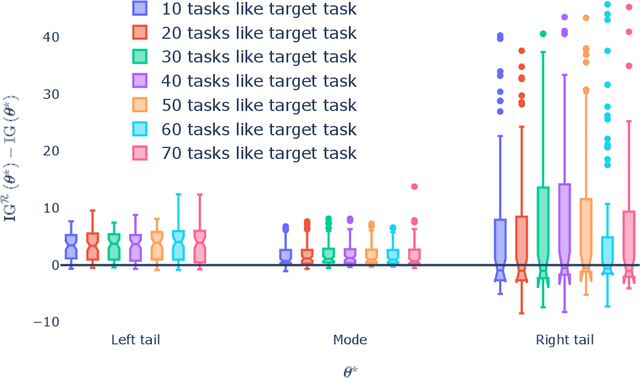
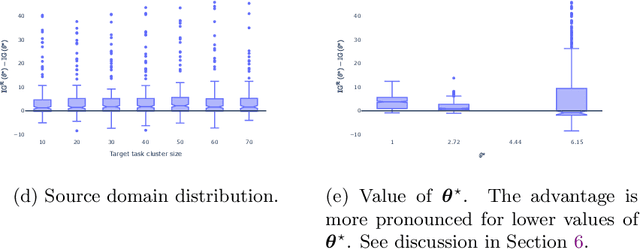
Abstract:Generalization outside the scope of one's training data requires leveraging prior knowledge about the effects that transfer, and the effects that don't, between different data sources. Bayesian transfer learning is a principled paradigm for specifying this knowledge, and refining it on the basis of data from the source (training) and target (prediction) tasks. We address the challenging transfer learning setting where the learner (i) cannot fine-tune in the target task, and (ii) does not know which source data points correspond to the same task (i.e., the data sources are unknown). We propose a proxy-informed robust method for probabilistic transfer learning (PROMPT), which provides a posterior predictive estimate tailored to the structure of the target task, without requiring the learner have access to any outcome information from the target task. Instead, PROMPT relies on the availability of proxy information. PROMPT uses the same proxy information for two purposes: (i) estimation of effects specific to the target task, and (ii) construction of a robust reweighting of the source data for estimation of effects that transfer between tasks. We provide theoretical results on the effect of this reweighting on the risk of negative transfer, and demonstrate application of PROMPT in two synthetic settings.
Heteroscedastic Preferential Bayesian Optimization with Informative Noise Distributions
May 23, 2024Abstract:Preferential Bayesian optimization (PBO) is a sample-efficient framework for learning human preferences between candidate designs. PBO classically relies on homoscedastic noise models to represent human aleatoric uncertainty. Yet, such noise fails to accurately capture the varying levels of human aleatoric uncertainty, particularly when the user possesses partial knowledge among different pairs of candidates. For instance, a chemist with solid expertise in glucose-related molecules may easily compare two compounds from that family while struggling to compare alcohol-related molecules. Currently, PBO overlooks this uncertainty during the search for a new candidate through the maximization of the acquisition function, consequently underestimating the risk associated with human uncertainty. To address this issue, we propose a heteroscedastic noise model to capture human aleatoric uncertainty. This model adaptively assigns noise levels based on the distance of a specific input to a predefined set of reliable inputs known as anchors provided by the human. Anchors encapsulate partial knowledge and offer insight into the comparative difficulty of evaluating different candidate pairs. Such a model can be seamlessly integrated into the acquisition function, thus leading to candidate design pairs that elegantly trade informativeness and ease of comparison for the human expert. We perform an extensive empirical evaluation of the proposed approach, demonstrating a consistent improvement over homoscedastic PBO.
Cost-aware learning of relevant contextual variables within Bayesian optimization
May 24, 2023
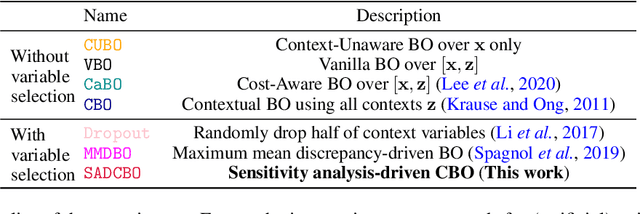
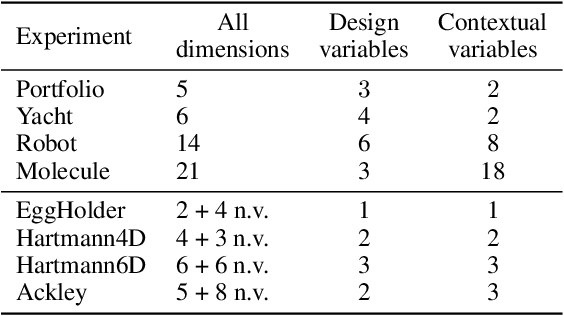

Abstract:Contextual Bayesian Optimization (CBO) is a powerful framework for optimizing black-box, expensive-to-evaluate functions with respect to design variables, while simultaneously efficiently integrating relevant contextual information regarding the environment, such as experimental conditions. However, in many practical scenarios, the relevance of contextual variables is not necessarily known beforehand. Moreover, the contextual variables can sometimes be optimized themselves, a setting that current CBO algorithms do not take into account. Optimizing contextual variables may be costly, which raises the question of determining a minimal relevant subset. In this paper, we frame this problem as a cost-aware model selection BO task and address it using a novel method, Sensitivity-Analysis-Driven Contextual BO (SADCBO). We learn the relevance of context variables by sensitivity analysis of the posterior surrogate model at specific input points, whilst minimizing the cost of optimization by leveraging recent developments on early stopping for BO. We empirically evaluate our proposed SADCBO against alternatives on synthetic experiments together with extensive ablation studies, and demonstrate a consistent improvement across examples.
Multi-Fidelity Bayesian Optimization with Unreliable Information Sources
Oct 25, 2022Abstract:Bayesian optimization (BO) is a powerful framework for optimizing black-box, expensive-to-evaluate functions. Over the past decade, many algorithms have been proposed to integrate cheaper, lower-fidelity approximations of the objective function into the optimization process, with the goal of converging towards the global optimum at a reduced cost. This task is generally referred to as multi-fidelity Bayesian optimization (MFBO). However, MFBO algorithms can lead to higher optimization costs than their vanilla BO counterparts, especially when the low-fidelity sources are poor approximations of the objective function, therefore defeating their purpose. To address this issue, we propose rMFBO (robust MFBO), a methodology to make any GP-based MFBO scheme robust to the addition of unreliable information sources. rMFBO comes with a theoretical guarantee that its performance can be bound to its vanilla BO analog, with high controllable probability. We demonstrate the effectiveness of the proposed methodology on a number of numerical benchmarks, outperforming earlier MFBO methods on unreliable sources. We expect rMFBO to be particularly useful to reliably include human experts with varying knowledge within BO processes.
Reactmine: a search algorithm for inferring chemical reaction networks from time series data
Sep 07, 2022

Abstract:Inferring chemical reaction networks (CRN) from time series data is a challenge encouraged by the growing availability of quantitative temporal data at the cellular level. This motivates the design of algorithms to infer the preponderant reactions between the molecular species observed in a given biochemical process, and help to build CRN model structure and kinetics. Existing ODE-based inference methods such as SINDy resort to least square regression combined with sparsity-enforcing penalization, such as Lasso. However, when the input time series are only available in wild type conditions in which all reactions are present, we observe that current methods fail to learn sparse models. Results: We present Reactmine, a CRN learning algorithm which enforces sparsity by inferring reactions in a sequential fashion within a search tree of bounded depth, ranking the inferred reaction candidates according to the variance of their kinetics, and re-optimizing the CRN kinetic parameters on the whole trace in a final pass to rank the inferred CRN candidates. We first evaluate its performance on simulation data from a benchmark of hidden CRNs, together with algorithmic hyperparameter sensitivity analyses, and then on two sets of real experimental data: one from protein fluorescence videomicroscopy of cell cycle and circadian clock markers, and one from biomedical measurements of systemic circadian biomarkers possibly acting on clock gene expression in peripheral organs. We show that Reactmine succeeds both on simulation data by retrieving hidden CRNs where SINDy fails, and on the two real datasets by inferring reactions in agreement with previous studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge