John Wu
PyHealth 2.0: A Comprehensive Open-Source Toolkit for Accessible and Reproducible Clinical Deep Learning
Jan 23, 2026Abstract:Difficulty replicating baselines, high computational costs, and required domain expertise create persistent barriers to clinical AI research. To address these challenges, we introduce PyHealth 2.0, an enhanced clinical deep learning toolkit that enables predictive modeling in as few as 7 lines of code. PyHealth 2.0 offers three key contributions: (1) a comprehensive toolkit addressing reproducibility and compatibility challenges by unifying 15+ datasets, 20+ clinical tasks, 25+ models, 5+ interpretability methods, and uncertainty quantification including conformal prediction within a single framework that supports diverse clinical data modalities - signals, imaging, and electronic health records - with translation of 5+ medical coding standards; (2) accessibility-focused design accommodating multimodal data and diverse computational resources with up to 39x faster processing and 20x lower memory usage, enabling work from 16GB laptops to production systems; and (3) an active open-source community of 400+ members lowering domain expertise barriers through extensive documentation, reproducible research contributions, and collaborations with academic health systems and industry partners, including multi-language support via RHealth. PyHealth 2.0 establishes an open-source foundation and community advancing accessible, reproducible healthcare AI. Available at pip install pyhealth.
Beyond Label Attention: Transparency in Language Models for Automated Medical Coding via Dictionary Learning
Oct 31, 2024



Abstract:Medical coding, the translation of unstructured clinical text into standardized medical codes, is a crucial but time-consuming healthcare practice. Though large language models (LLM) could automate the coding process and improve the efficiency of such tasks, interpretability remains paramount for maintaining patient trust. Current efforts in interpretability of medical coding applications rely heavily on label attention mechanisms, which often leads to the highlighting of extraneous tokens irrelevant to the ICD code. To facilitate accurate interpretability in medical language models, this paper leverages dictionary learning that can efficiently extract sparsely activated representations from dense language model embeddings in superposition. Compared with common label attention mechanisms, our model goes beyond token-level representations by building an interpretable dictionary which enhances the mechanistic-based explanations for each ICD code prediction, even when the highlighted tokens are medically irrelevant. We show that dictionary features can steer model behavior, elucidate the hidden meanings of upwards of 90% of medically irrelevant tokens, and are human interpretable.
DILA: Dictionary Label Attention for Mechanistic Interpretability in High-dimensional Multi-label Medical Coding Prediction
Sep 16, 2024



Abstract:Predicting high-dimensional or extreme multilabels, such as in medical coding, requires both accuracy and interpretability. Existing works often rely on local interpretability methods, failing to provide comprehensive explanations of the overall mechanism behind each label prediction within a multilabel set. We propose a mechanistic interpretability module called DIctionary Label Attention (\method) that disentangles uninterpretable dense embeddings into a sparse embedding space, where each nonzero element (a dictionary feature) represents a globally learned medical concept. Through human evaluations, we show that our sparse embeddings are more human understandable than its dense counterparts by at least 50 percent. Our automated dictionary feature identification pipeline, leveraging large language models (LLMs), uncovers thousands of learned medical concepts by examining and summarizing the highest activating tokens for each dictionary feature. We represent the relationships between dictionary features and medical codes through a sparse interpretable matrix, enhancing the mechanistic and global understanding of the model's predictions while maintaining competitive performance and scalability without extensive human annotation.
If LLM Is the Wizard, Then Code Is the Wand: A Survey on How Code Empowers Large Language Models to Serve as Intelligent Agents
Jan 08, 2024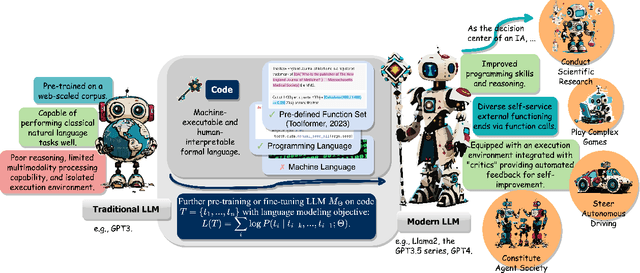
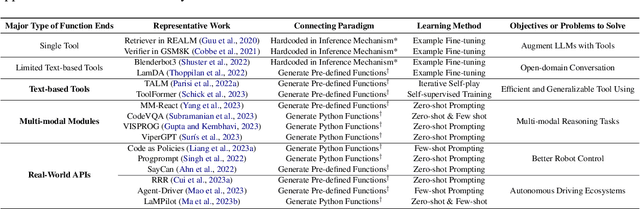
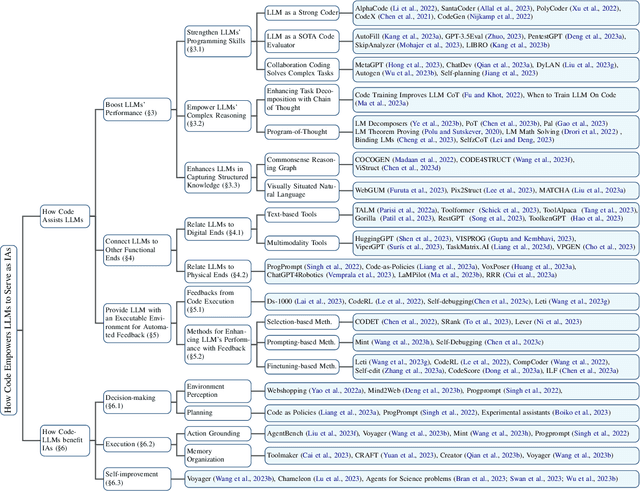

Abstract:The prominent large language models (LLMs) of today differ from past language models not only in size, but also in the fact that they are trained on a combination of natural language and formal language (code). As a medium between humans and computers, code translates high-level goals into executable steps, featuring standard syntax, logical consistency, abstraction, and modularity. In this survey, we present an overview of the various benefits of integrating code into LLMs' training data. Specifically, beyond enhancing LLMs in code generation, we observe that these unique properties of code help (i) unlock the reasoning ability of LLMs, enabling their applications to a range of more complex natural language tasks; (ii) steer LLMs to produce structured and precise intermediate steps, which can then be connected to external execution ends through function calls; and (iii) take advantage of code compilation and execution environment, which also provides diverse feedback for model improvement. In addition, we trace how these profound capabilities of LLMs, brought by code, have led to their emergence as intelligent agents (IAs) in situations where the ability to understand instructions, decompose goals, plan and execute actions, and refine from feedback are crucial to their success on downstream tasks. Finally, we present several key challenges and future directions of empowering LLMs with code.
A Case for Dataset Specific Profiling
Aug 01, 2022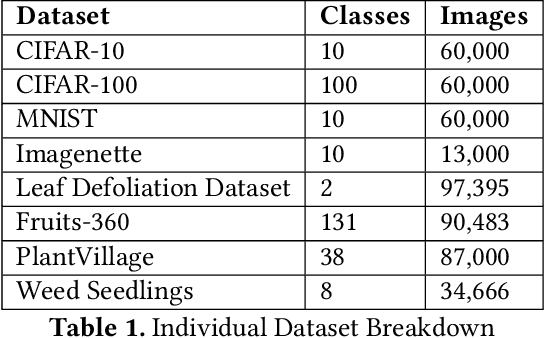
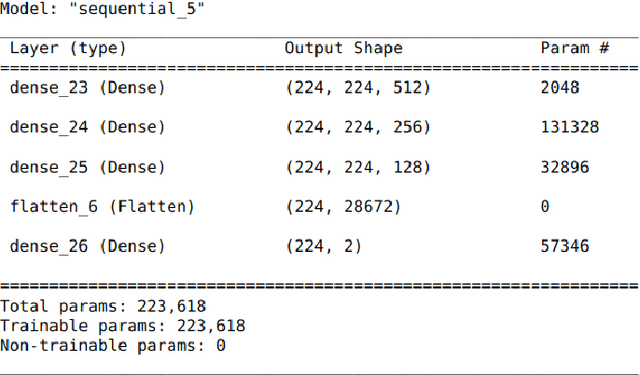
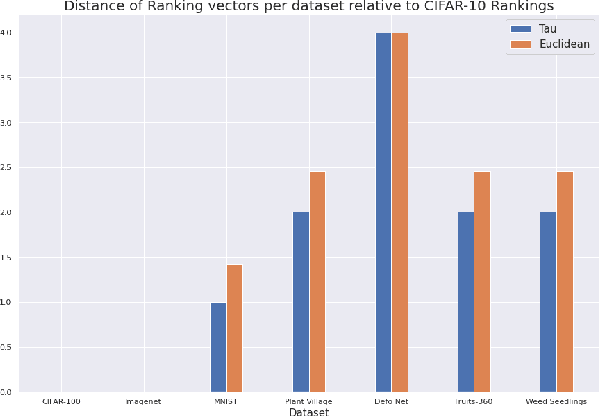
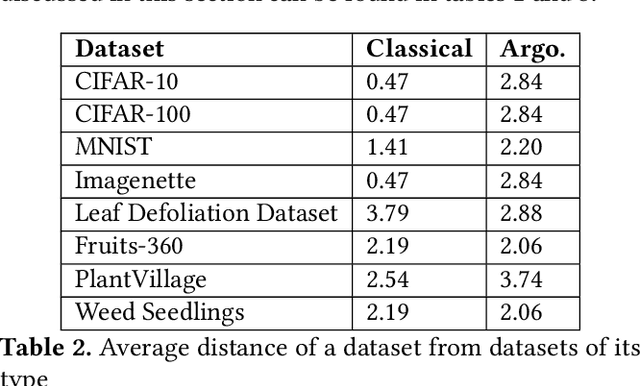
Abstract:Data-driven science is an emerging paradigm where scientific discoveries depend on the execution of computational AI models against rich, discipline-specific datasets. With modern machine learning frameworks, anyone can develop and execute computational models that reveal concepts hidden in the data that could enable scientific applications. For important and widely used datasets, computing the performance of every computational model that can run against a dataset is cost prohibitive in terms of cloud resources. Benchmarking approaches used in practice use representative datasets to infer performance without actually executing models. While practicable, these approaches limit extensive dataset profiling to a few datasets and introduce bias that favors models suited for representative datasets. As a result, each dataset's unique characteristics are left unexplored and subpar models are selected based on inference from generalized datasets. This necessitates a new paradigm that introduces dataset profiling into the model selection process. To demonstrate the need for dataset-specific profiling, we answer two questions:(1) Can scientific datasets significantly permute the rank order of computational models compared to widely used representative datasets? (2) If so, could lightweight model execution improve benchmarking accuracy? Taken together, the answers to these questions lay the foundation for a new dataset-aware benchmarking paradigm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge