Jinglin Zhao
DTN: Deep Multiple Task-specific Feature Interactions Network for Multi-Task Recommendation
Aug 21, 2024



Abstract:Neural-based multi-task learning (MTL) has been successfully applied to many recommendation applications. However, these MTL models (e.g., MMoE, PLE) did not consider feature interaction during the optimization, which is crucial for capturing complex high-order features and has been widely used in ranking models for real-world recommender systems. Moreover, through feature importance analysis across various tasks in MTL, we have observed an interesting divergence phenomenon that the same feature can have significantly different importance across different tasks in MTL. To address these issues, we propose Deep Multiple Task-specific Feature Interactions Network (DTN) with a novel model structure design. DTN introduces multiple diversified task-specific feature interaction methods and task-sensitive network in MTL networks, enabling the model to learn task-specific diversified feature interaction representations, which improves the efficiency of joint representation learning in a general setup. We applied DTN to our company's real-world E-commerce recommendation dataset, which consisted of over 6.3 billion samples, the results demonstrated that DTN significantly outperformed state-of-the-art MTL models. Moreover, during online evaluation of DTN in a large-scale E-commerce recommender system, we observed a 3.28% in clicks, a 3.10% increase in orders and a 2.70% increase in GMV (Gross Merchandise Value) compared to the state-of-the-art MTL models. Finally, extensive offline experiments conducted on public benchmark datasets demonstrate that DTN can be applied to various scenarios beyond recommendations, enhancing the performance of ranking models.
A Topological Nomenclature for 3D Shape Analysis in Connectomics
Sep 27, 2019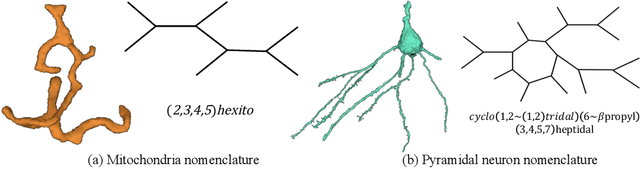
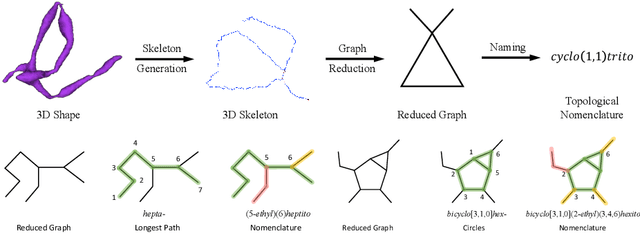
Abstract:An essential task in nano-scale connectomics is the morphology analysis of neurons and organelles like mitochondria to shed light on their biological properties. However, these biological objects often have tangled parts or complex branching patterns, which makes it hard to abstract, categorize, and manipulate their morphology. Here we propose a topological nomenclature to name these objects like chemical compounds for neuroscience analysis. To this end, we convert the volumetric representation into the topology-preserving reduced graph, develop nomenclature rules for pyramidal neurons and mitochondria from the reduced graph, and learn the feature embedding for shape manipulation. In ablation studies, we show that the proposed reduced graph extraction method yield graphs better in accord with the perception of experts. On 3D shape retrieval and decomposition tasks, we show that the encoded topological nomenclature features achieve better results than state-of-the-art shape descriptors. To advance neuroscience, we will release a 3D mesh dataset of mitochondria and pyramidal neurons reconstructed from a 100{\mu}m cube electron microscopy (EM) volume. Code is publicly available at https://github.com/donglaiw/ibexHelper.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge