Jin Su
U-Fold: Dynamic Intent-Aware Context Folding for User-Centric Agents
Jan 26, 2026Abstract:Large language model (LLM)-based agents have been successfully deployed in many tool-augmented settings, but their scalability is fundamentally constrained by context length. Existing context-folding methods mitigate this issue by summarizing past interactions, yet they are typically designed for single-query or single-intent scenarios. In more realistic user-centric dialogues, we identify two major failure modes: (i) they irreversibly discard fine-grained constraints and intermediate facts that are crucial for later decisions, and (ii) their summaries fail to track evolving user intent, leading to omissions and erroneous actions. To address these limitations, we propose U-Fold, a dynamic context-folding framework tailored to user-centric tasks. U-Fold retains the full user--agent dialogue and tool-call history but, at each turn, uses two core components to produce an intent-aware, evolving dialogue summary and a compact, task-relevant tool log. Extensive experiments on $τ$-bench, $τ^2$-bench, VitaBench, and harder context-inflated settings show that U-Fold consistently outperforms ReAct (achieving a 71.4% win rate in long-context settings) and prior folding baselines (with improvements of up to 27.0%), particularly on long, noisy, multi-turn tasks. Our study demonstrates that U-Fold is a promising step toward transferring context-management techniques from single-query benchmarks to realistic user-centric applications.
Protein 3D Graph Structure Learning for Robust Structure-based Protein Property Prediction
Oct 19, 2023Abstract:Protein structure-based property prediction has emerged as a promising approach for various biological tasks, such as protein function prediction and sub-cellular location estimation. The existing methods highly rely on experimental protein structure data and fail in scenarios where these data are unavailable. Predicted protein structures from AI tools (e.g., AlphaFold2) were utilized as alternatives. However, we observed that current practices, which simply employ accurately predicted structures during inference, suffer from notable degradation in prediction accuracy. While similar phenomena have been extensively studied in general fields (e.g., Computer Vision) as model robustness, their impact on protein property prediction remains unexplored. In this paper, we first investigate the reason behind the performance decrease when utilizing predicted structures, attributing it to the structure embedding bias from the perspective of structure representation learning. To study this problem, we identify a Protein 3D Graph Structure Learning Problem for Robust Protein Property Prediction (PGSL-RP3), collect benchmark datasets, and present a protein Structure embedding Alignment Optimization framework (SAO) to mitigate the problem of structure embedding bias between the predicted and experimental protein structures. Extensive experiments have shown that our framework is model-agnostic and effective in improving the property prediction of both predicted structures and experimental structures. The benchmark datasets and codes will be released to benefit the community.
Exploring evolution-based & -free protein language models as protein function predictors
Jun 14, 2022
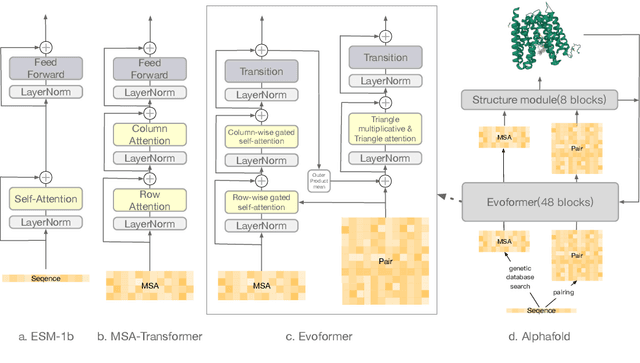

Abstract:Large-scale Protein Language Models (PLMs) have improved performance in protein prediction tasks, ranging from 3D structure prediction to various function predictions. In particular, AlphaFold, a ground-breaking AI system, could potentially reshape structural biology. However, the utility of the PLM module in AlphaFold, Evoformer, has not been explored beyond structure prediction. In this paper, we investigate the representation ability of three popular PLMs: ESM-1b (single sequence), MSA-Transformer (multiple sequence alignment) and Evoformer (structural), with a special focus on Evoformer. Specifically, we aim to answer the following key questions: (i) Does the Evoformer trained as part of AlphaFold produce representations amenable to predicting protein function? (ii) If yes, can Evoformer replace ESM-1b and MSA-Transformer? (iii) How much do these PLMs rely on evolution-related protein data? In this regard, are they complementary to each other? We compare these models by empirical study along with new insights and conclusions. Finally, we release code and datasets for reproducibility.
Multi-granularity Textual Adversarial Attack with Behavior Cloning
Sep 09, 2021



Abstract:Recently, the textual adversarial attack models become increasingly popular due to their successful in estimating the robustness of NLP models. However, existing works have obvious deficiencies. (1) They usually consider only a single granularity of modification strategies (e.g. word-level or sentence-level), which is insufficient to explore the holistic textual space for generation; (2) They need to query victim models hundreds of times to make a successful attack, which is highly inefficient in practice. To address such problems, in this paper we propose MAYA, a Multi-grAnularitY Attack model to effectively generate high-quality adversarial samples with fewer queries to victim models. Furthermore, we propose a reinforcement-learning based method to train a multi-granularity attack agent through behavior cloning with the expert knowledge from our MAYA algorithm to further reduce the query times. Additionally, we also adapt the agent to attack black-box models that only output labels without confidence scores. We conduct comprehensive experiments to evaluate our attack models by attacking BiLSTM, BERT and RoBERTa in two different black-box attack settings and three benchmark datasets. Experimental results show that our models achieve overall better attacking performance and produce more fluent and grammatical adversarial samples compared to baseline models. Besides, our adversarial attack agent significantly reduces the query times in both attack settings. Our codes are released at https://github.com/Yangyi-Chen/MAYA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge