Jicong Zhang
Sparsity- and Hybridity-Inspired Visual Parameter-Efficient Fine-Tuning for Medical Diagnosis
May 28, 2024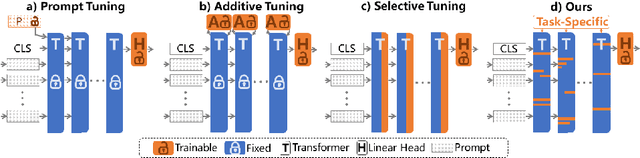
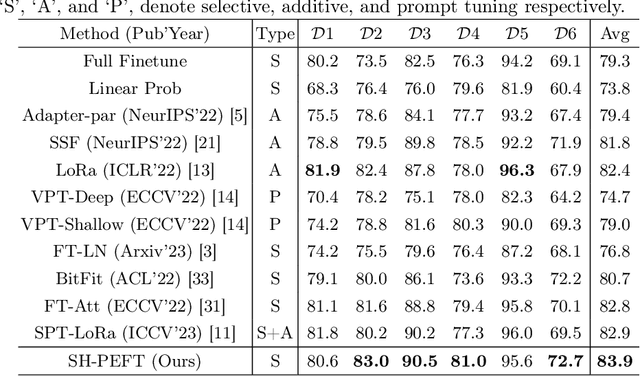
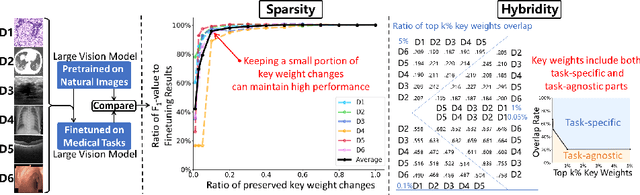
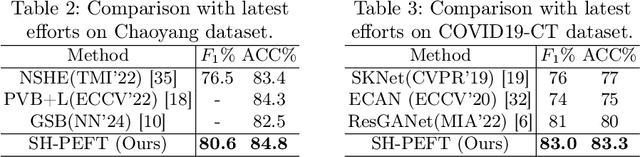
Abstract:The success of Large Vision Models (LVMs) is accompanied by vast data volumes, which are prohibitively expensive in medical diagnosis.To address this, recent efforts exploit Parameter-Efficient Fine-Tuning (PEFT), which trains a small number of weights while freezing the rest.However, they typically assign trainable weights to the same positions in LVMs in a heuristic manner, regardless of task differences, making them suboptimal for professional applications like medical diagnosis.To address this, we statistically reveal the nature of sparsity and hybridity during diagnostic-targeted fine-tuning, i.e., a small portion of key weights significantly impacts performance, and these key weights are hybrid, including both task-specific and task-agnostic parts.Based on this, we propose a novel Sparsity- and Hybridity-inspired Parameter Efficient Fine-Tuning (SH-PEFT).It selects and trains a small portion of weights based on their importance, which is innovatively estimated by hybridizing both task-specific and task-agnostic strategies.Validated on six medical datasets of different modalities, we demonstrate that SH-PEFT achieves state-of-the-art performance in transferring LVMs to medical diagnosis in terms of accuracy. By tuning around 0.01% number of weights, it outperforms full model fine-tuning.Moreover, SH-PEFT also achieves comparable performance to other models deliberately optimized for specific medical tasks.Extensive experiments demonstrate the effectiveness of each design and reveal that large model transfer holds great potential in medical diagnosis.
Learning Large Margin Sparse Embeddings for Open Set Medical Diagnosis
Jul 21, 2023Abstract:Fueled by deep learning, computer-aided diagnosis achieves huge advances. However, out of controlled lab environments, algorithms could face multiple challenges. Open set recognition (OSR), as an important one, states that categories unseen in training could appear in testing. In medical fields, it could derive from incompletely collected training datasets and the constantly emerging new or rare diseases. OSR requires an algorithm to not only correctly classify known classes, but also recognize unknown classes and forward them to experts for further diagnosis. To tackle OSR, we assume that known classes could densely occupy small parts of the embedding space and the remaining sparse regions could be recognized as unknowns. Following it, we propose Open Margin Cosine Loss (OMCL) unifying two mechanisms. The former, called Margin Loss with Adaptive Scale (MLAS), introduces angular margin for reinforcing intra-class compactness and inter-class separability, together with an adaptive scaling factor to strengthen the generalization capacity. The latter, called Open-Space Suppression (OSS), opens the classifier by recognizing sparse embedding space as unknowns using proposed feature space descriptors. Besides, since medical OSR is still a nascent field, two publicly available benchmark datasets are proposed for comparison. Extensive ablation studies and feature visualization demonstrate the effectiveness of each design. Compared with state-of-the-art methods, MLAS achieves superior performances, measured by ACC, AUROC, and OSCR.
Learning with Limited Annotations: A Survey on Deep Semi-Supervised Learning for Medical Image Segmentation
Aug 13, 2022
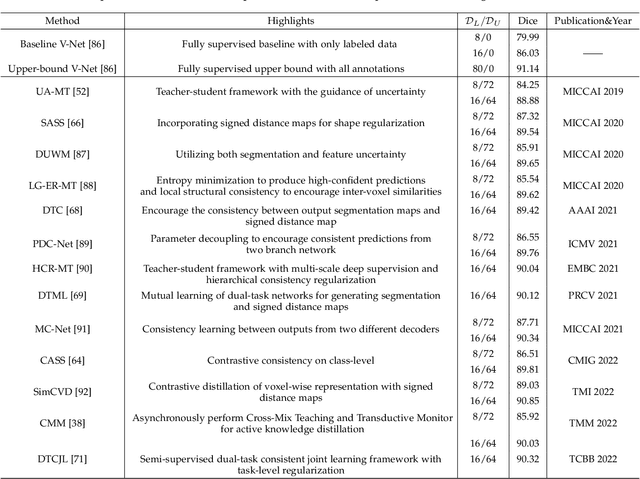
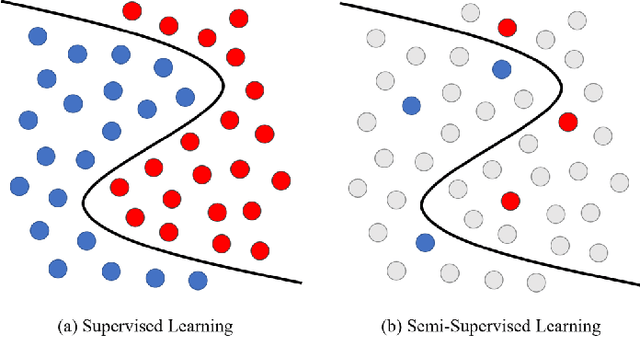
Abstract:Medical image segmentation is a fundamental and critical step in many image-guided clinical approaches. Recent success of deep learning-based segmentation methods usually relies on a large amount of labeled data, which is particularly difficult and costly to obtain especially in the medical imaging domain where only experts can provide reliable and accurate annotations. Semi-supervised learning has emerged as an appealing strategy and been widely applied to medical image segmentation tasks to train deep models with limited annotations. In this paper, we present a comprehensive review of recently proposed semi-supervised learning methods for medical image segmentation and summarized both the technical novelties and empirical results. Furthermore, we analyze and discuss the limitations and several unsolved problems of existing approaches. We hope this review could inspire the research community to explore solutions for this challenge and further promote the developments in medical image segmentation field.
Parallel Network with Channel Attention and Post-Processing for Carotid Arteries Vulnerable Plaque Segmentation in Ultrasound Images
Apr 18, 2022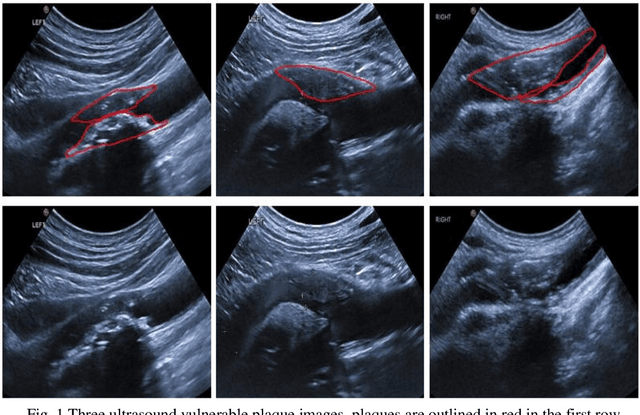
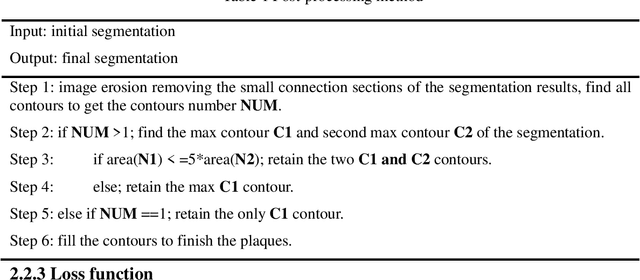
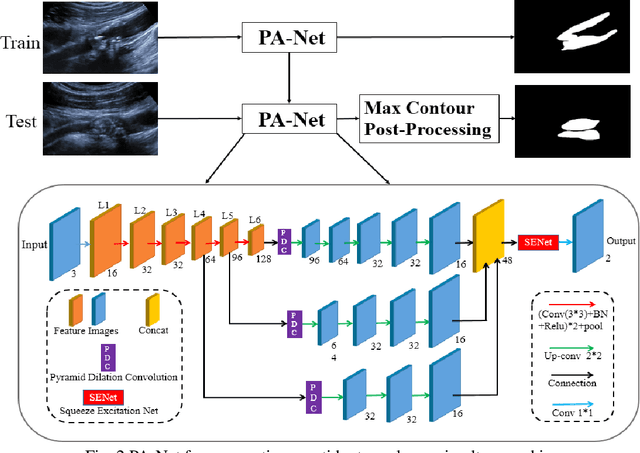
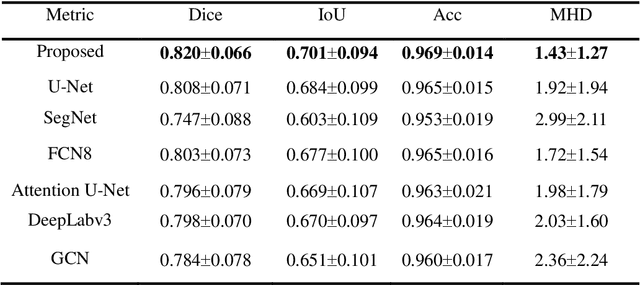
Abstract:Carotid arteries vulnerable plaques are a crucial factor in the screening of atherosclerosis by ultrasound technique. However, the plaques are contaminated by various noises such as artifact, speckle noise, and manual segmentation may be time-consuming. This paper proposes an automatic convolutional neural network (CNN) method for plaque segmentation in carotid ultrasound images using a small dataset. First, a parallel network with three independent scale decoders is utilized as our base segmentation network, pyramid dilation convolutions are used to enlarge receptive fields in the three segmentation sub-networks. Subsequently, the three decoders are merged to be rectified in channels by SENet. Thirdly, in test stage, the initially segmented plaque is refined by the max contour morphology post-processing to obtain the final plaque. Moreover, three loss function Dice loss, SSIM loss and cross-entropy loss are compared to segment plaques. Test results show that the proposed method with dice loss function yields a Dice value of 0.820, an IoU of 0.701, Acc of 0.969, and modified Hausdorff distance (MHD) of 1.43 for 30 vulnerable cases of plaques, it outperforms some of the conventional CNN-based methods on these metrics. Additionally, we apply an ablation experiment to show the validity of each proposed module. Our study provides some reference for similar researches and may be useful in actual applications for plaque segmentation of ultrasound carotid arteries.
Uncertainty-Guided Mutual Consistency Learning for Semi-Supervised Medical Image Segmentation
Dec 05, 2021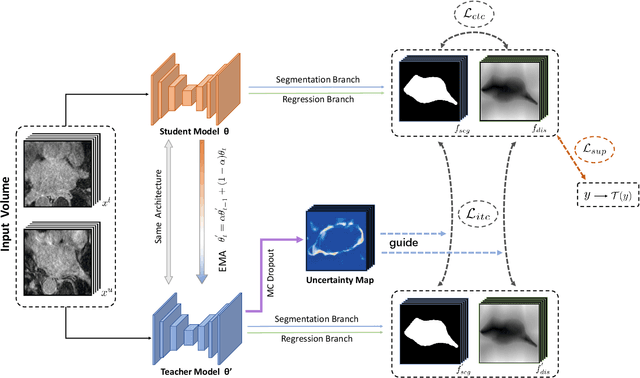
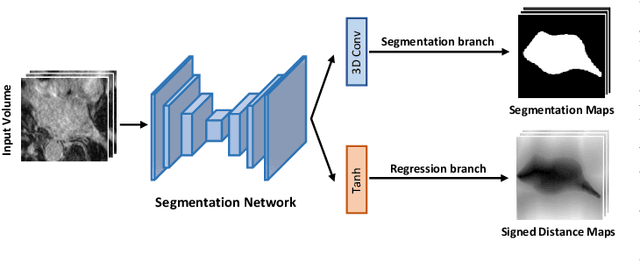
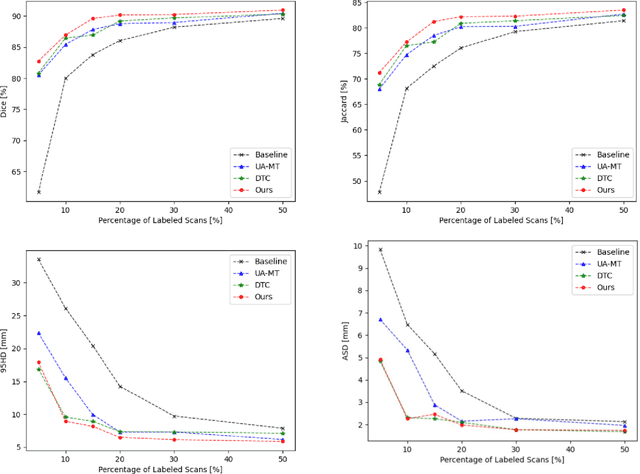
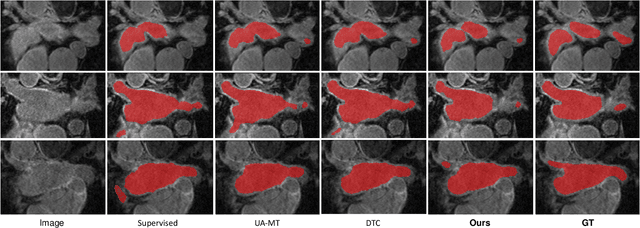
Abstract:Medical image segmentation is a fundamental and critical step in many clinical approaches. Semi-supervised learning has been widely applied to medical image segmentation tasks since it alleviates the heavy burden of acquiring expert-examined annotations and takes the advantage of unlabeled data which is much easier to acquire. Although consistency learning has been proven to be an effective approach by enforcing an invariance of predictions under different distributions, existing approaches cannot make full use of region-level shape constraint and boundary-level distance information from unlabeled data. In this paper, we propose a novel uncertainty-guided mutual consistency learning framework to effectively exploit unlabeled data by integrating intra-task consistency learning from up-to-date predictions for self-ensembling and cross-task consistency learning from task-level regularization to exploit geometric shape information. The framework is guided by the estimated segmentation uncertainty of models to select out relatively certain predictions for consistency learning, so as to effectively exploit more reliable information from unlabeled data. We extensively validate our proposed method on two publicly available benchmark datasets: Left Atrium Segmentation (LA) dataset and Brain Tumor Segmentation (BraTS) dataset. Experimental results demonstrate that our method achieves performance gains by leveraging unlabeled data and outperforms existing semi-supervised segmentation methods.
Parameter Decoupling Strategy for Semi-supervised 3D Left Atrium Segmentation
Sep 20, 2021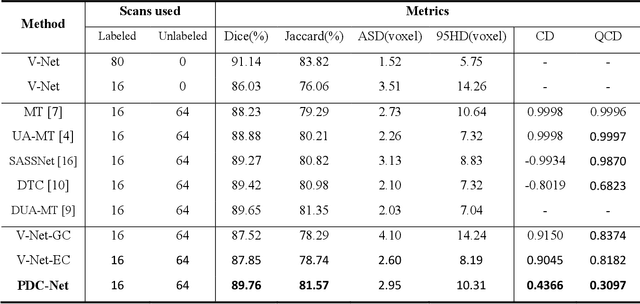
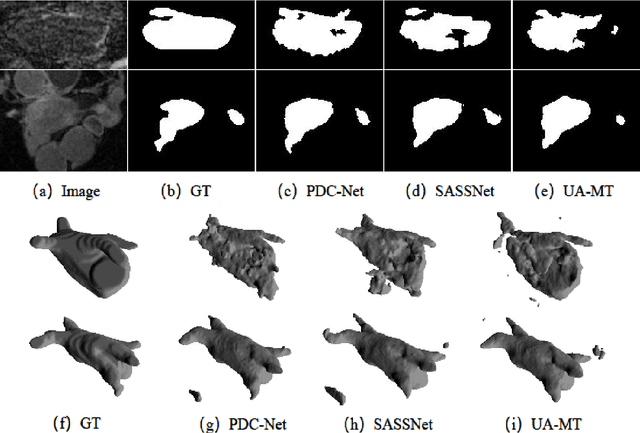
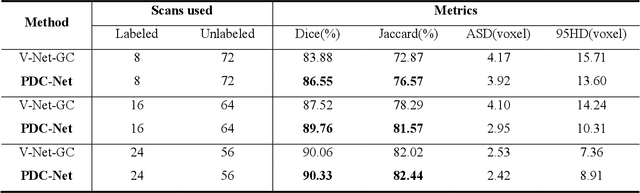
Abstract:Consistency training has proven to be an advanced semi-supervised framework and achieved promising results in medical image segmentation tasks through enforcing an invariance of the predictions over different views of the inputs. However, with the iterative updating of model parameters, the models would tend to reach a coupled state and eventually lose the ability to exploit unlabeled data. To address the issue, we present a novel semi-supervised segmentation model based on parameter decoupling strategy to encourage consistent predictions from diverse views. Specifically, we first adopt a two-branch network to simultaneously produce predictions for each image. During the training process, we decouple the two prediction branch parameters by quadratic cosine distance to construct different views in latent space. Based on this, the feature extractor is constrained to encourage the consistency of probability maps generated by classifiers under diversified features. In the overall training process, the parameters of feature extractor and classifiers are updated alternately by consistency regularization operation and decoupling operation to gradually improve the generalization performance of the model. Our method has achieved a competitive result over the state-of-the-art semi-supervised methods on the Atrial Segmentation Challenge dataset, demonstrating the effectiveness of our framework. Code is available at https://github.com/BX0903/PDC.
Dual-Task Mutual Learning for Semi-Supervised Medical Image Segmentation
Mar 08, 2021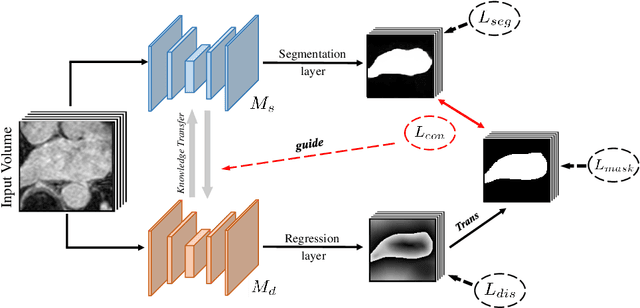
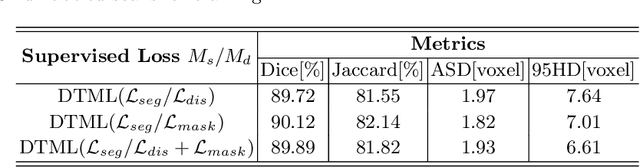
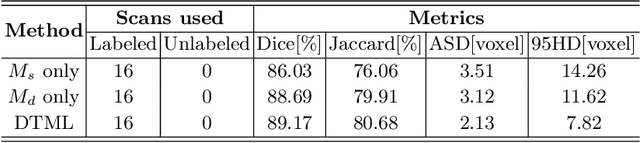
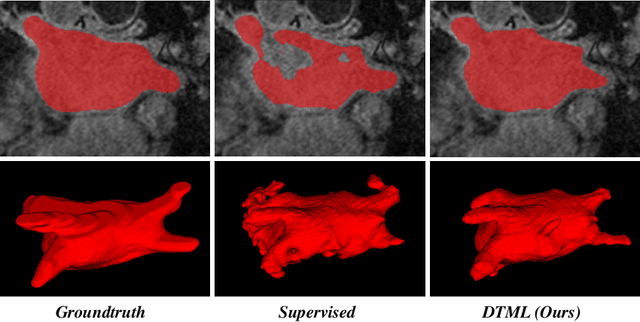
Abstract:The success of deep learning methods in medical image segmentation tasks usually requires a large amount of labeled data. However, obtaining reliable annotations is expensive and time-consuming. Semi-supervised learning has attracted much attention in medical image segmentation by taking the advantage of unlabeled data which is much easier to acquire. In this paper, we propose a novel dual-task mutual learning framework for semi-supervised medical image segmentation. Our framework can be formulated as an integration of two individual segmentation networks based on two tasks: learning region-based shape constraint and learning boundary-based surface mismatch. Different from the one-way transfer between teacher and student networks, an ensemble of dual-task students can learn collaboratively and implicitly explore useful knowledge from each other during the training process. By jointly learning the segmentation probability maps and signed distance maps of targets, our framework can enforce the geometric shape constraint and learn more reliable information. Experimental results demonstrate that our method achieves performance gains by leveraging unlabeled data and outperforms the state-of-the-art semi-supervised segmentation methods.
Exploiting Shared Knowledge from Non-COVID Lesions for Annotation-Efficient COVID-19 CT Lung Infection Segmentation
Dec 31, 2020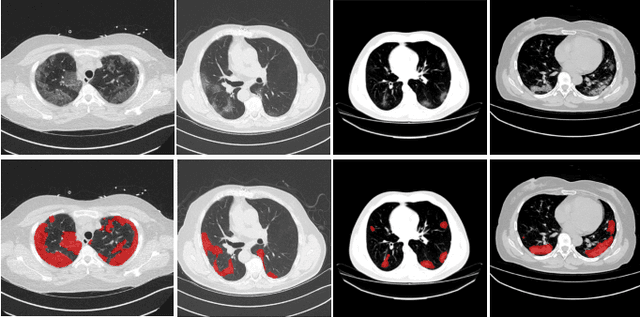
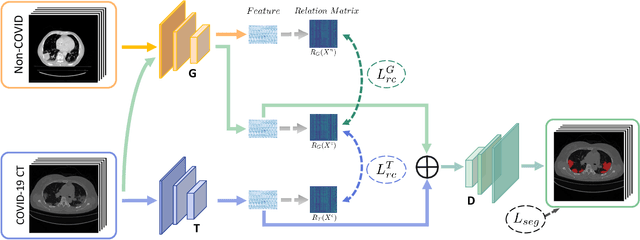
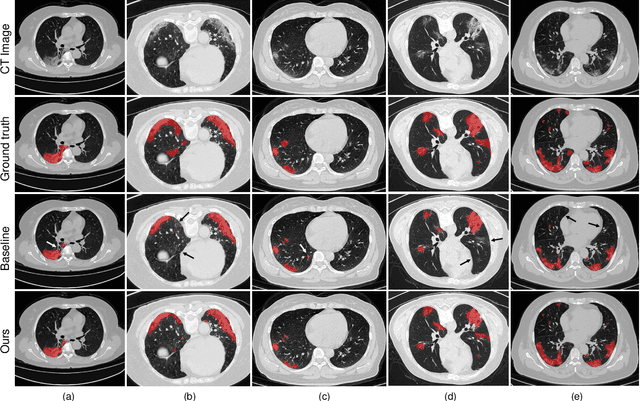
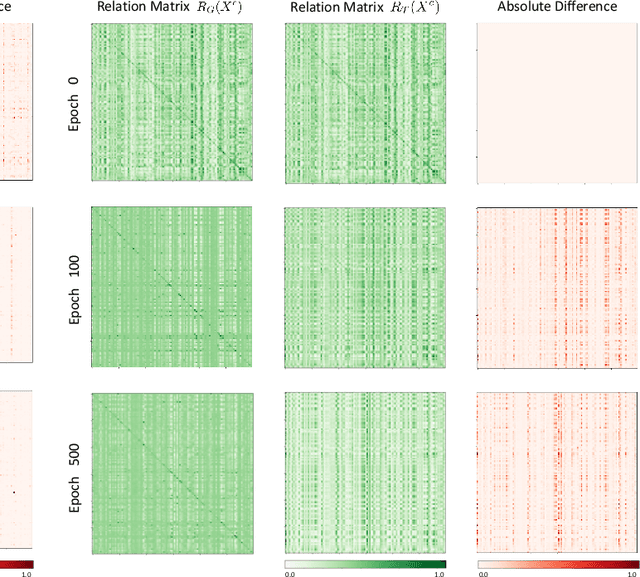
Abstract:The novel Coronavirus disease (COVID-19) is a highly contagious virus and has spread all over the world, posing an extremely serious threat to all countries. Automatic lung infection segmentation from computed tomography (CT) plays an important role in the quantitative analysis of COVID-19. However, the major challenge lies in the inadequacy of annotated COVID-19 datasets. Currently, there are several public non-COVID lung lesion segmentation datasets, providing the potential for generalizing useful information to the related COVID-19 segmentation task. In this paper, we propose a novel relation-driven collaborative learning model for annotation-efficient COVID-19 CT lung infection segmentation. The network consists of encoders with the same architecture and a shared decoder. The general encoder is adopted to capture general lung lesion features based on multiple non-COVID lesions, while the target encoder is adopted to focus on task-specific features of COVID-19 infections. Features extracted from the two parallel encoders are concatenated for the subsequent decoder part. To thoroughly exploit shared knowledge between COVID and non-COVID lesions, we develop a collaborative learning scheme to regularize the relation consistency between extracted features of given input. Other than existing consistency-based methods that simply enforce the consistency of individual predictions, our method enforces the consistency of feature relation among samples, encouraging the model to explore semantic information from both COVID-19 and non-COVID cases. Extensive experiments on one public COVID-19 dataset and two public non-COVID datasets show that our method achieves superior segmentation performance compared with existing methods in the absence of sufficient high-quality COVID-19 annotations.
Exploring Efficient Volumetric Medical Image Segmentation Using 2.5D Method: An Empirical Study
Oct 13, 2020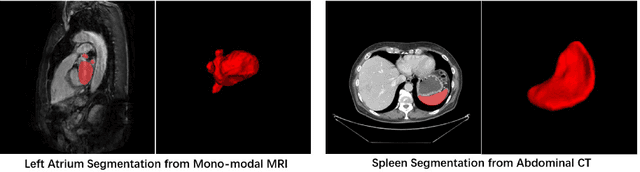
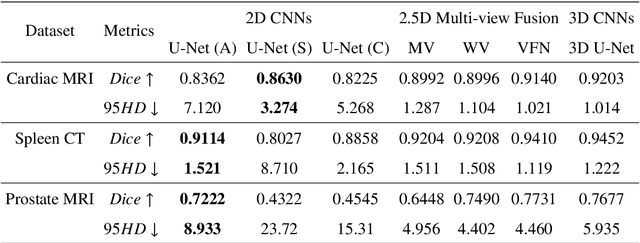
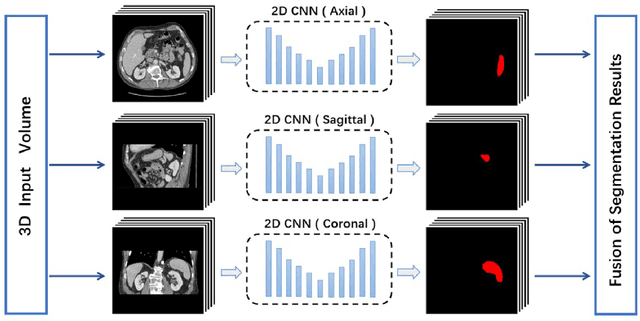
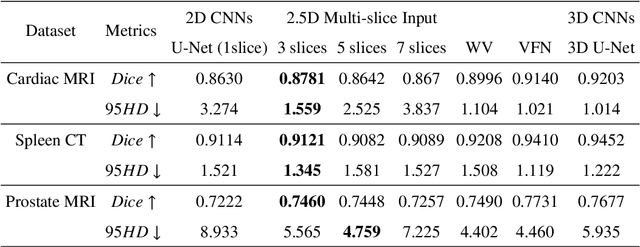
Abstract:With the unprecedented developments in deep learning, many methods are proposed and have achieved great success for medical image segmentation. However, unlike segmentation of natural images, most medical images such as MRI and CT are volumetric data. In order to make full use of volumetric information, 3D CNNs are widely used. However, 3D CNNs suffer from higher inference time and computation cost, which hinders their further clinical applications. Additionally, with the increased number of parameters, the risk of overfitting is higher, especially for medical images where data and annotations are expensive to acquire. To issue this problem, many 2.5D segmentation methods have been proposed to make use of volumetric spatial information with less computation cost. Despite these works lead to improvements on a variety of segmentation tasks, to the best of our knowledge, there has not previously been a large-scale empirical comparison of these methods. In this paper, we aim to present a review of the latest developments of 2.5D methods for volumetric medical image segmentation. Additionally, to compare the performance and effectiveness of these methods, we provide an empirical study of these methods on three representative segmentation tasks involving different modalities and targets. Our experimental results highlight that 3D CNNs may not always be the best choice. Besides, although all these 2.5D methods can bring performance gains to 2D baseline, not all the methods hold the benefits on different datasets. We hope the results and conclusions of our study will prove useful for the community on exploring and developing efficient volumetric medical image segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge